Analysis of Journal Effect
Adam H. Sparks
2024-08-07
Source:vignettes/f_analysis_journal.Rmd
f_analysis_journal.RmdSet-up Workspace
Load libraries used and setting the ggplot2 theme for the document.
library("brms")
library("bayestestR")
library("bayesplot")
library("ggplot2")
library("here")
library("pander")
library("report")
library("tidyr")
library("Reproducibility.in.Plant.Pathology")
options(mc.cores = parallel::detectCores())
theme_set(theme_classic())Notes
2024-08-07
While preparing a talk about this paper and compendium, typos and
other minor errors were corrected in these analyses. However, there is
a bug in
{report::report()} that prevents the report from being generated for a
{brms} model, it also duplicates
text if it does generate it. Therefore, code to generate the reports
are commented out and the original report objects are
maintained along with the original model objects that were reported in
the paper in the “Save the Model Objects” section.
Introduction
This vignette documents the analysis of the data gathered from surveying 21 journals and 450 articles in the field of plant pathology for their openness and reproducibility and the effect that the journal it was published in had on that score.
Journal Effect Models
Computational Methods Availability
Test the effect that the journal’s effect on the available of
computational methods. Set up a brms model with journal
(abbreviation) as the fixed effect and
assignee as a random effect. Test the effect that the
journal may have on the availability of data. Base level is set to
Phytopathology, the journal that will be used as the intercept
in the following model.
# import data
rrpp <- import_notes()
# relevel factors for analysis, use Phytopathology as basis for analysis, highest IF
rrpp <-
within(rrpp,
abbreviation <-
relevel(abbreviation, ref = "Phytopathology"))
rrpp <- drop_na(rrpp, comp_mthds_avail)
m_f1 <-
brm(
formula = comp_mthds_avail ~ abbreviation +
(1 | assignee),
data = rrpp,
seed = 27,
prior = priors,
family = cumulative(),
iter = 10000,
control = list(adapt_delta = 0.99)
)
#> Compiling Stan program...
#> Start sampling
summary(m_f1)
#> Family: cumulative
#> Links: mu = logit; disc = identity
#> Formula: comp_mthds_avail ~ abbreviation + (1 | assignee)
#> Data: rrpp (Number of observations: 440)
#> Draws: 4 chains, each with iter = 10000; warmup = 5000; thin = 1;
#> total post-warmup draws = 20000
#>
#> Multilevel Hyperparameters:
#> ~assignee (Number of levels: 5)
#> Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
#> sd(Intercept) 6.23 2.71 2.80 13.09 1.00 8426 12548
#>
#> Regression Coefficients:
#> Estimate Est.Error l-95% CI u-95% CI Rhat
#> Intercept[1] 0.17 0.79 -1.38 1.73 1.00
#> Intercept[2] 0.55 0.79 -0.98 2.11 1.00
#> abbreviationAustralasPlantPath -0.14 0.94 -2.03 1.67 1.00
#> abbreviationCanJPlantPathol -0.21 0.92 -2.05 1.53 1.00
#> abbreviationCropProt -0.24 0.89 -2.02 1.45 1.00
#> abbreviationEurJPlantPathol -0.19 0.90 -2.01 1.54 1.00
#> abbreviationForestPathol -0.21 0.91 -2.04 1.53 1.00
#> abbreviationJPhytopathol -0.35 0.85 -2.08 1.26 1.00
#> abbreviationJPlantPathol -0.23 0.91 -2.06 1.53 1.00
#> abbreviationMolPlantMicroIn 0.47 0.84 -1.23 2.07 1.00
#> abbreviationMolPlantPathol -0.27 0.88 -2.06 1.38 1.00
#> abbreviationNematology -0.22 0.90 -2.03 1.49 1.00
#> abbreviationPhysiolMolPlantP -0.24 0.90 -2.05 1.46 1.00
#> abbreviationPhytoparasitica -0.26 0.88 -2.04 1.40 1.00
#> abbreviationPhytopatholMediterr -0.22 0.90 -2.03 1.51 1.00
#> abbreviationPlantDis -0.20 0.92 -2.03 1.56 1.00
#> abbreviationPlantHealthProgress -0.17 0.93 -2.05 1.61 1.00
#> abbreviationPlantPathol -0.26 0.89 -2.08 1.42 1.00
#> abbreviationRevMexFitopatol -0.24 0.91 -2.06 1.48 1.00
#> abbreviationTropPlantPathol 0.64 0.87 -1.10 2.31 1.00
#> abbreviationVirolJ -0.15 0.94 -2.04 1.65 1.00
#> Bulk_ESS Tail_ESS
#> Intercept[1] 23241 13755
#> Intercept[2] 25727 15591
#> abbreviationAustralasPlantPath 35310 14110
#> abbreviationCanJPlantPathol 34229 15128
#> abbreviationCropProt 37291 13326
#> abbreviationEurJPlantPathol 34264 14619
#> abbreviationForestPathol 36177 13900
#> abbreviationJPhytopathol 35051 14055
#> abbreviationJPlantPathol 36255 13822
#> abbreviationMolPlantMicroIn 37075 14611
#> abbreviationMolPlantPathol 34996 14100
#> abbreviationNematology 36274 14558
#> abbreviationPhysiolMolPlantP 36007 14484
#> abbreviationPhytoparasitica 36708 14625
#> abbreviationPhytopatholMediterr 34960 14291
#> abbreviationPlantDis 35575 13880
#> abbreviationPlantHealthProgress 37032 14610
#> abbreviationPlantPathol 34671 14255
#> abbreviationRevMexFitopatol 36299 13418
#> abbreviationTropPlantPathol 35965 15242
#> abbreviationVirolJ 40178 13424
#>
#> Further Distributional Parameters:
#> Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
#> disc 1.00 0.00 1.00 1.00 NA NA NA
#>
#> Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
#> and Tail_ESS are effective sample size measures, and Rhat is the potential
#> scale reduction factor on split chains (at convergence, Rhat = 1).
plot(m_f1)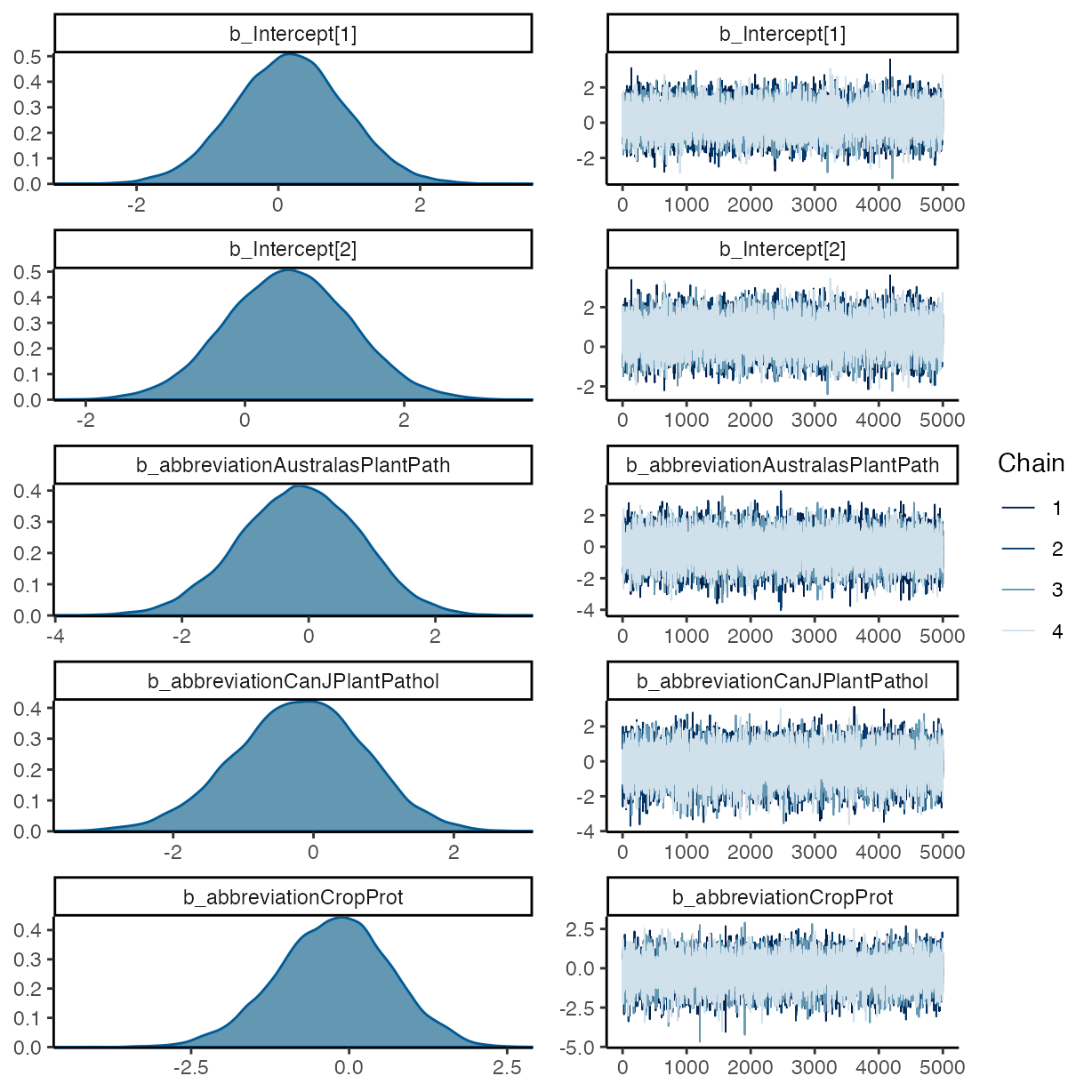
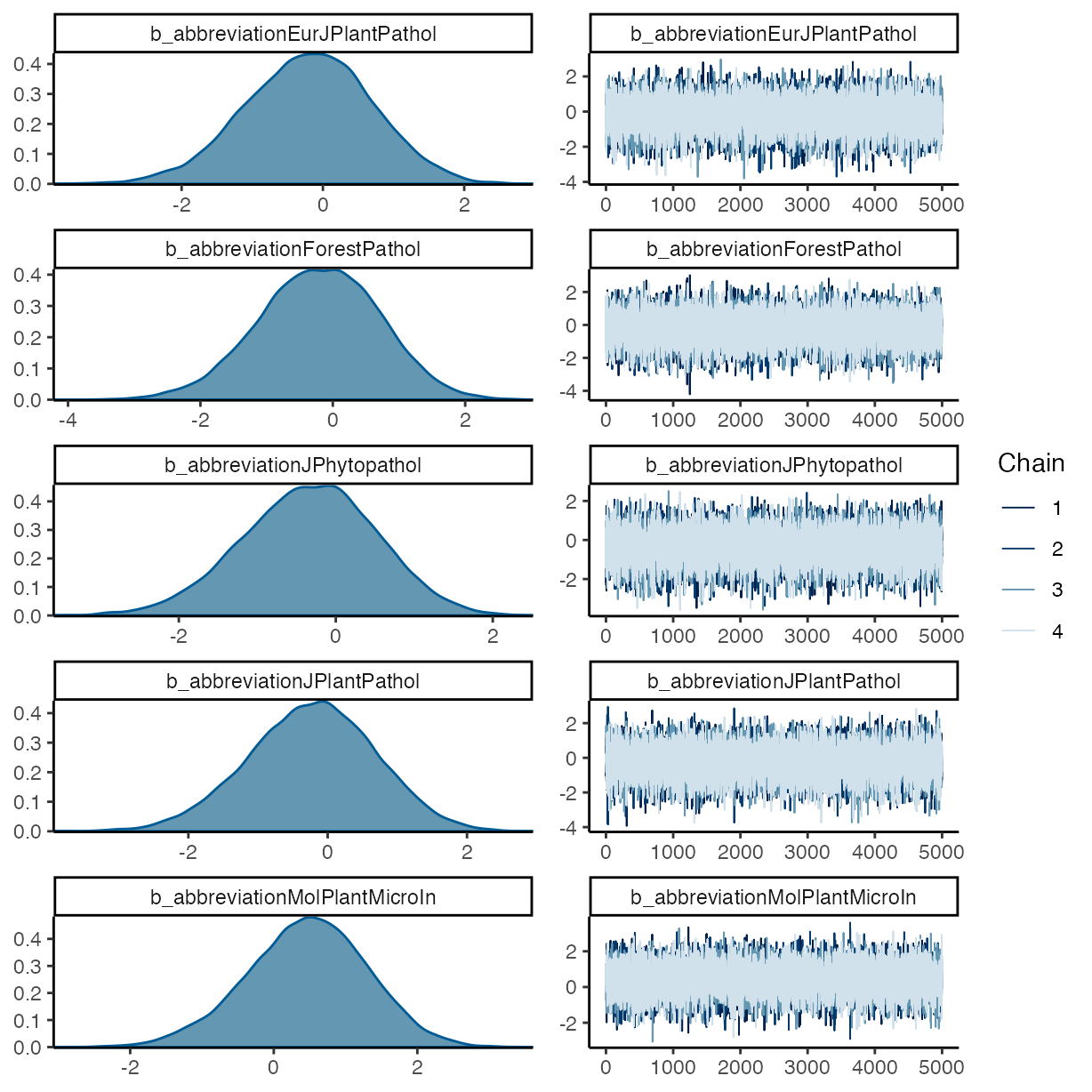
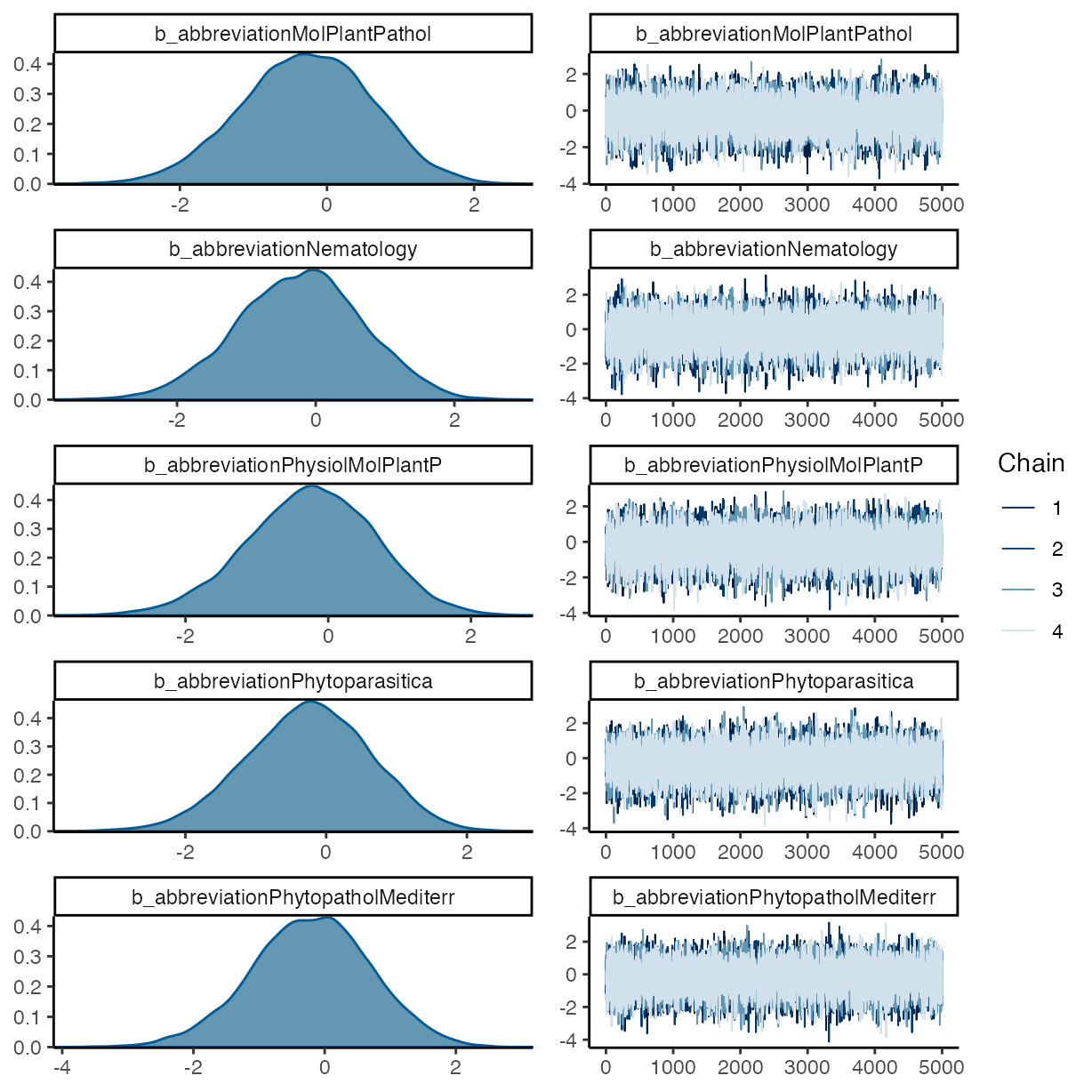
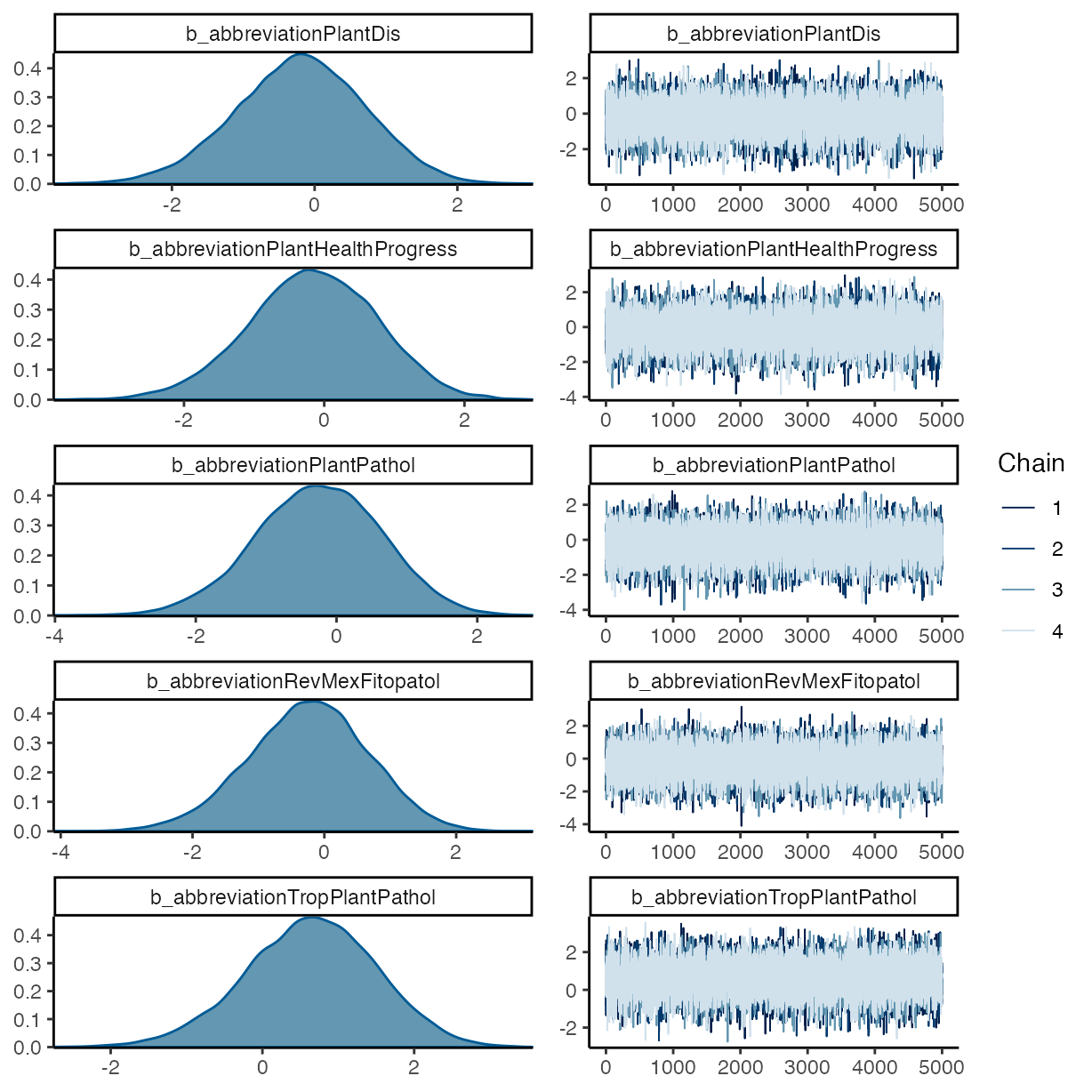
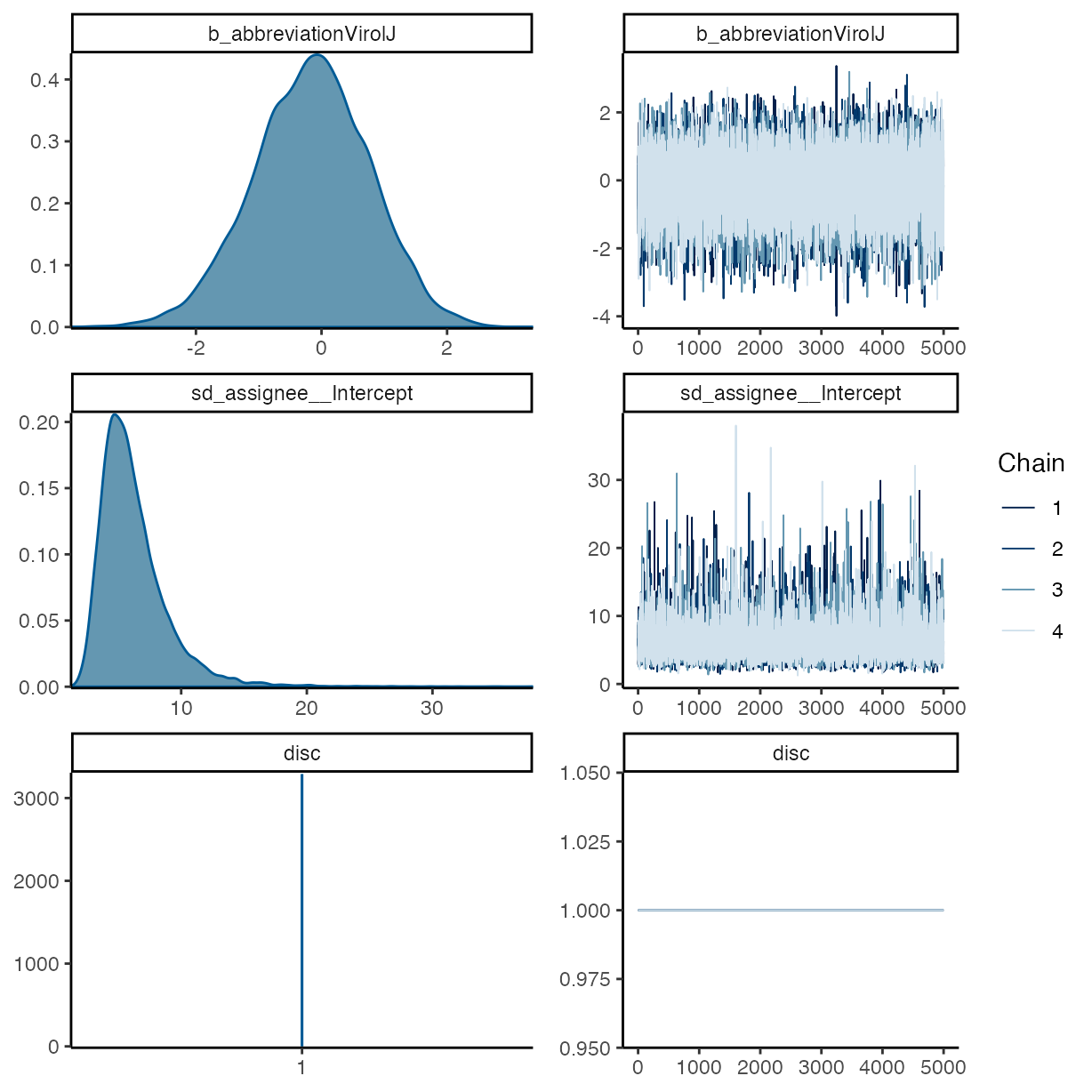
pp_check(m_f1, ndraws = 50, type = "bars")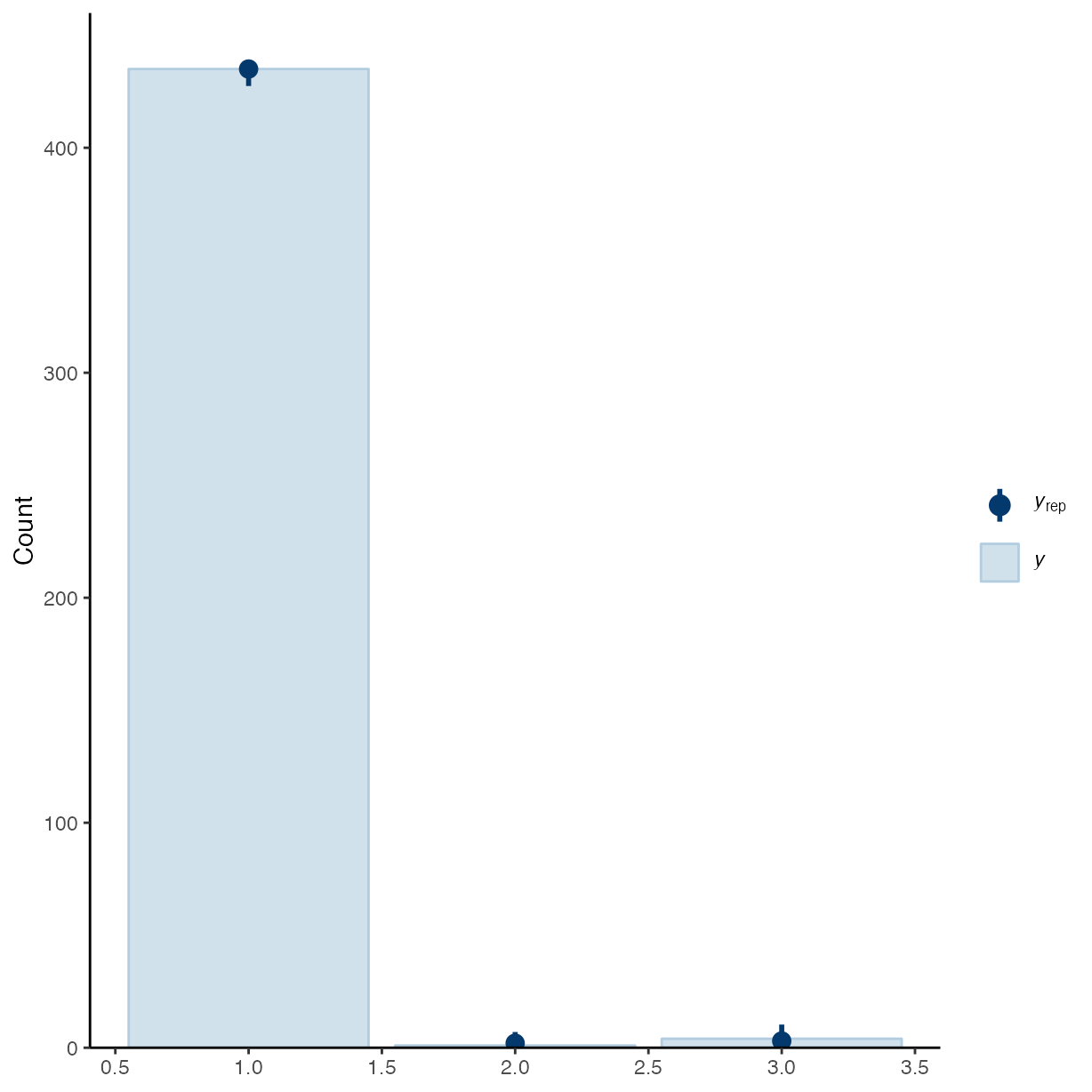
plot(equivalence_test(m_f1))
#> Picking joint bandwidth of 0.0983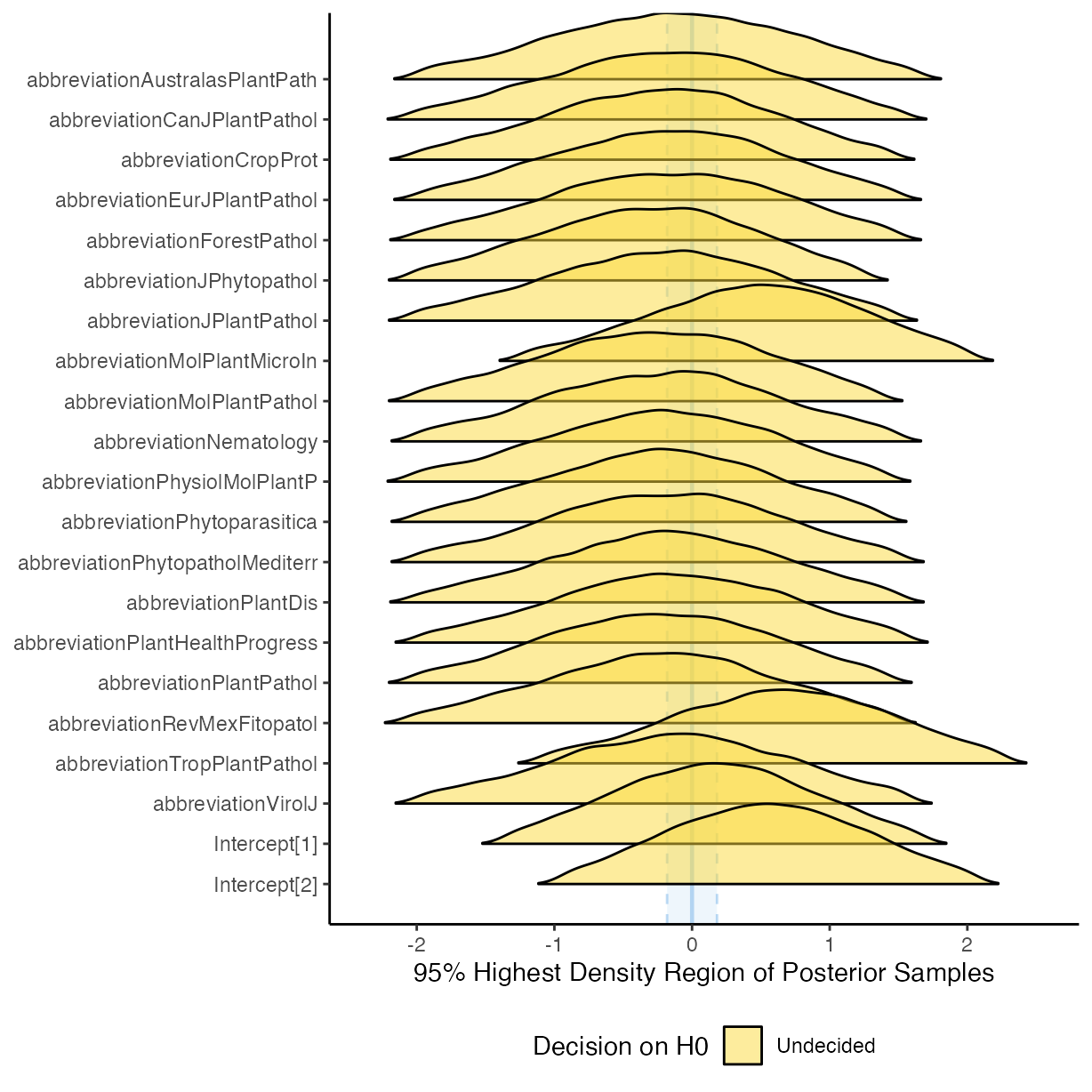
# pander(m_f1_report <- report(m_f1))
# m_f1_es <- report_effectsize(m_f1)Data Availability
Test the effect that the journal may have on the availability of data. Base level is set to Phytopathology, the journal that will be used as the intercept in the following model.
# reimport data due to changes in previous model dropping NA values
rrpp <- import_notes()
rrpp <-
within(rrpp,
abbreviation <-
relevel(abbreviation, ref = "Phytopathology"))
rrpp <- drop_na(rrpp, data_avail)
m_f2 <-
brm(
formula = data_avail ~ abbreviation +
(1 | assignee),
data = rrpp,
seed = 27,
prior = priors,
family = cumulative(),
iter = 10000,
control = list(adapt_delta = 0.99)
)
#> Compiling Stan program...
#> Start sampling
summary(m_f2)
#> Family: cumulative
#> Links: mu = logit; disc = identity
#> Formula: data_avail ~ abbreviation + (1 | assignee)
#> Data: rrpp (Number of observations: 448)
#> Draws: 4 chains, each with iter = 10000; warmup = 5000; thin = 1;
#> total post-warmup draws = 20000
#>
#> Multilevel Hyperparameters:
#> ~assignee (Number of levels: 5)
#> Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
#> sd(Intercept) 2.44 1.37 0.47 5.76 1.00 3406 3047
#>
#> Regression Coefficients:
#> Estimate Est.Error l-95% CI u-95% CI Rhat
#> Intercept[1] 0.41 0.69 -0.95 1.72 1.00
#> Intercept[2] 0.68 0.69 -0.68 1.99 1.00
#> Intercept[3] 1.15 0.69 -0.22 2.46 1.00
#> abbreviationAustralasPlantPath 0.60 0.64 -0.70 1.83 1.00
#> abbreviationCanJPlantPathol 0.29 0.54 -0.78 1.31 1.00
#> abbreviationCropProt -1.26 0.70 -2.72 0.03 1.00
#> abbreviationEurJPlantPathol 0.06 0.57 -1.11 1.13 1.00
#> abbreviationForestPathol -0.11 0.56 -1.26 0.96 1.00
#> abbreviationJPhytopathol -0.27 0.47 -1.23 0.63 1.00
#> abbreviationJPlantPathol 0.14 0.54 -0.97 1.16 1.00
#> abbreviationMolPlantMicroIn 0.53 0.49 -0.47 1.46 1.00
#> abbreviationMolPlantPathol 0.93 0.42 0.07 1.73 1.00
#> abbreviationNematology -0.22 0.63 -1.52 0.96 1.00
#> abbreviationPhysiolMolPlantP 0.58 0.47 -0.35 1.46 1.00
#> abbreviationPhytoparasitica -0.49 0.59 -1.70 0.62 1.00
#> abbreviationPhytopatholMediterr 1.67 0.46 0.77 2.57 1.00
#> abbreviationPlantDis -1.29 0.69 -2.73 -0.02 1.00
#> abbreviationPlantHealthProgress -0.62 0.67 -1.99 0.63 1.00
#> abbreviationPlantPathol -0.11 0.50 -1.12 0.83 1.00
#> abbreviationRevMexFitopatol -1.17 0.70 -2.62 0.14 1.00
#> abbreviationTropPlantPathol 0.34 0.55 -0.75 1.38 1.00
#> abbreviationVirolJ 0.87 0.44 -0.01 1.73 1.00
#> Bulk_ESS Tail_ESS
#> Intercept[1] 4898 5597
#> Intercept[2] 4979 5662
#> Intercept[3] 5124 5927
#> abbreviationAustralasPlantPath 25235 15027
#> abbreviationCanJPlantPathol 24072 13686
#> abbreviationCropProt 27703 13730
#> abbreviationEurJPlantPathol 23893 14456
#> abbreviationForestPathol 24456 13857
#> abbreviationJPhytopathol 20919 13601
#> abbreviationJPlantPathol 26010 14417
#> abbreviationMolPlantMicroIn 22190 13851
#> abbreviationMolPlantPathol 20272 15550
#> abbreviationNematology 26596 14005
#> abbreviationPhysiolMolPlantP 20574 14192
#> abbreviationPhytoparasitica 23165 13869
#> abbreviationPhytopatholMediterr 20010 14913
#> abbreviationPlantDis 27463 13483
#> abbreviationPlantHealthProgress 26598 13873
#> abbreviationPlantPathol 21843 14727
#> abbreviationRevMexFitopatol 28469 13739
#> abbreviationTropPlantPathol 23756 14317
#> abbreviationVirolJ 20571 14149
#>
#> Further Distributional Parameters:
#> Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
#> disc 1.00 0.00 1.00 1.00 NA NA NA
#>
#> Draws were sampled using sampling(NUTS). For each parameter, Bulk_ESS
#> and Tail_ESS are effective sample size measures, and Rhat is the potential
#> scale reduction factor on split chains (at convergence, Rhat = 1).
plot(m_f2)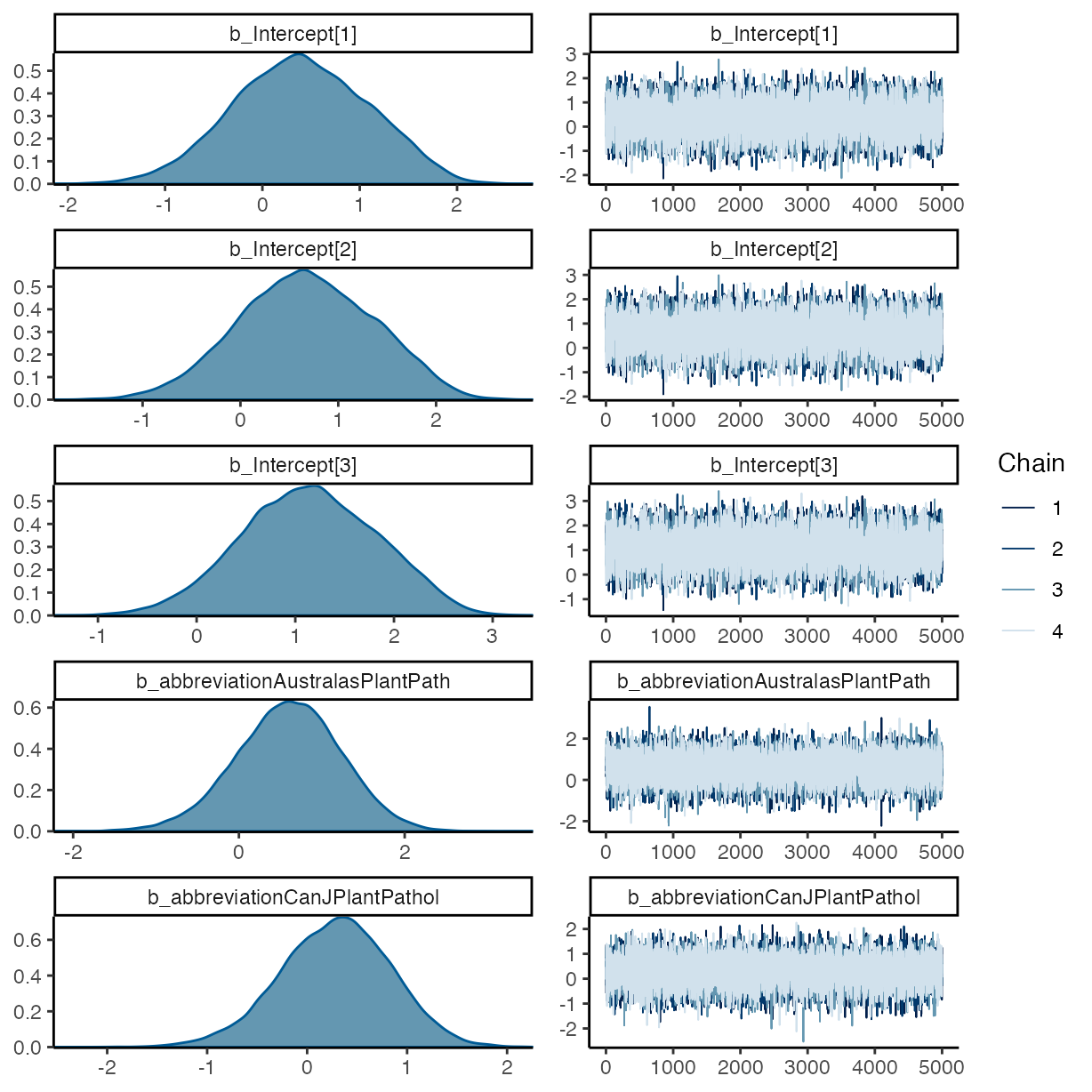
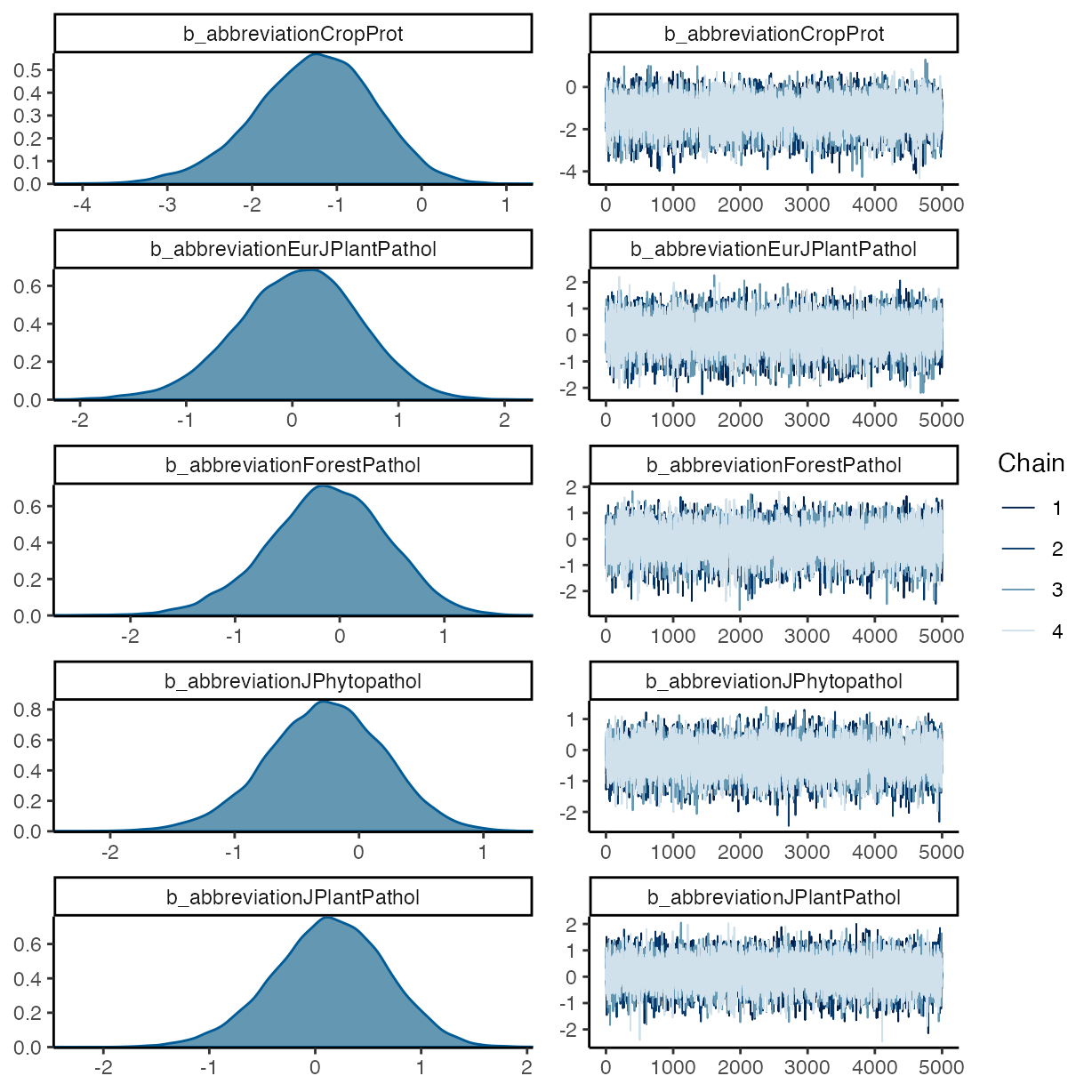
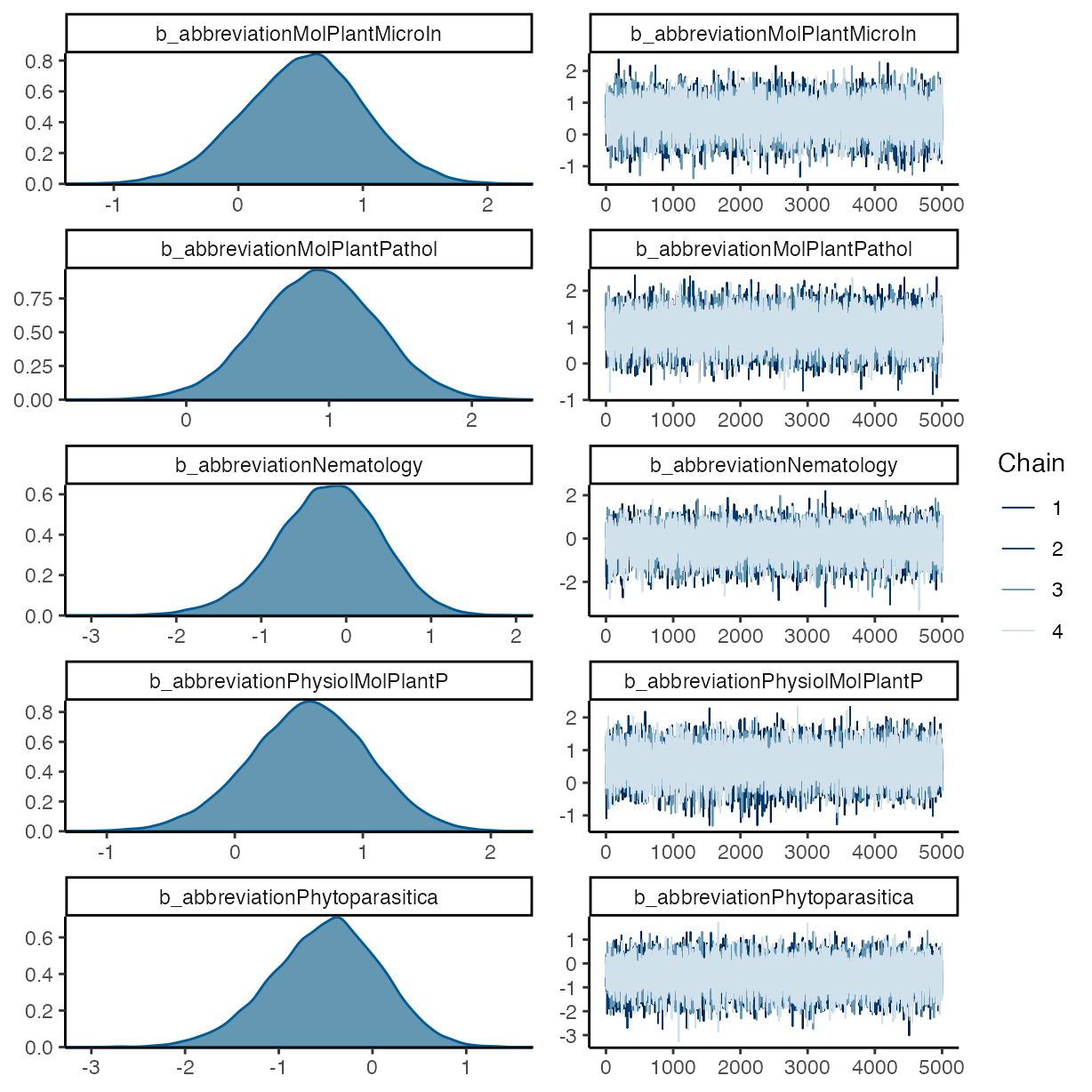
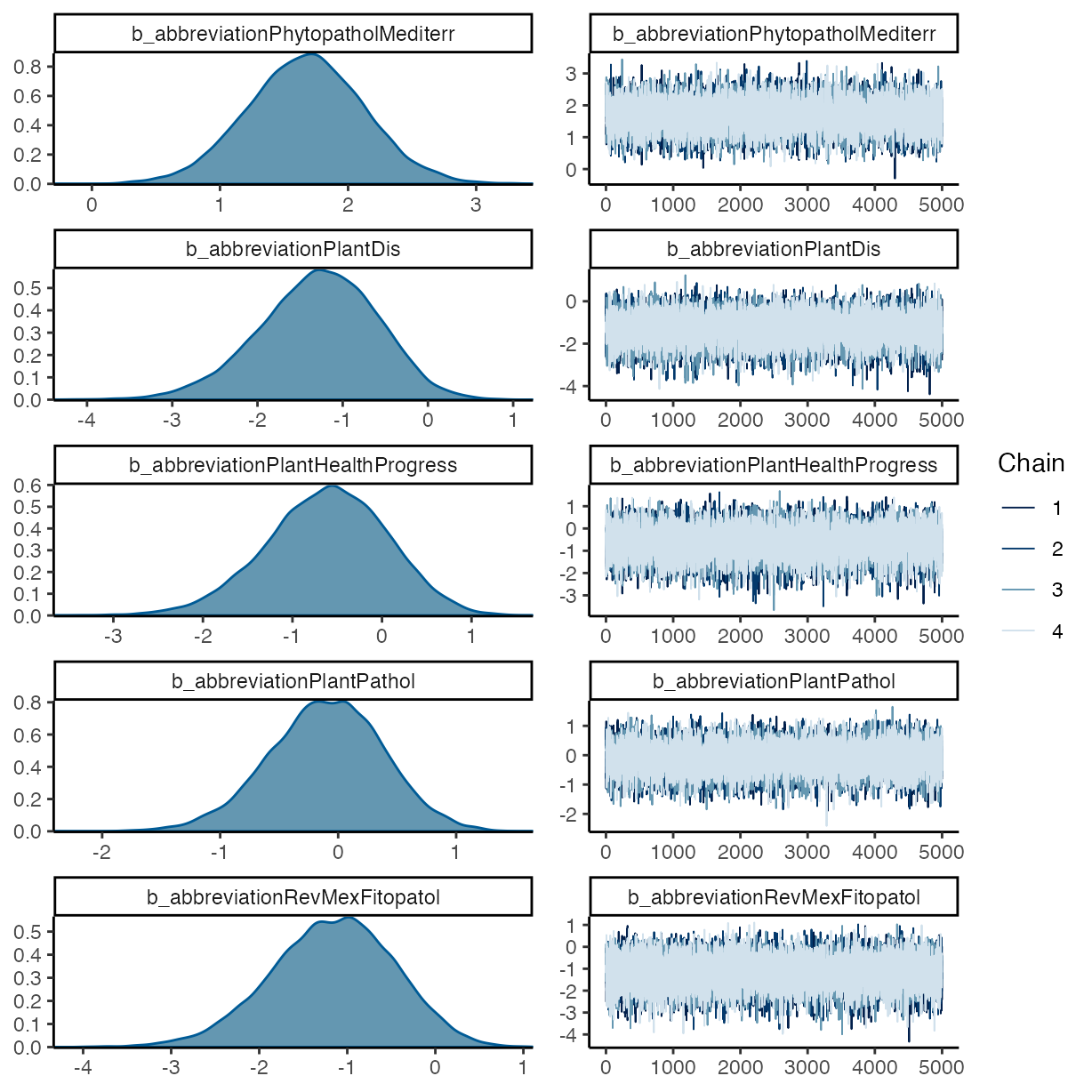
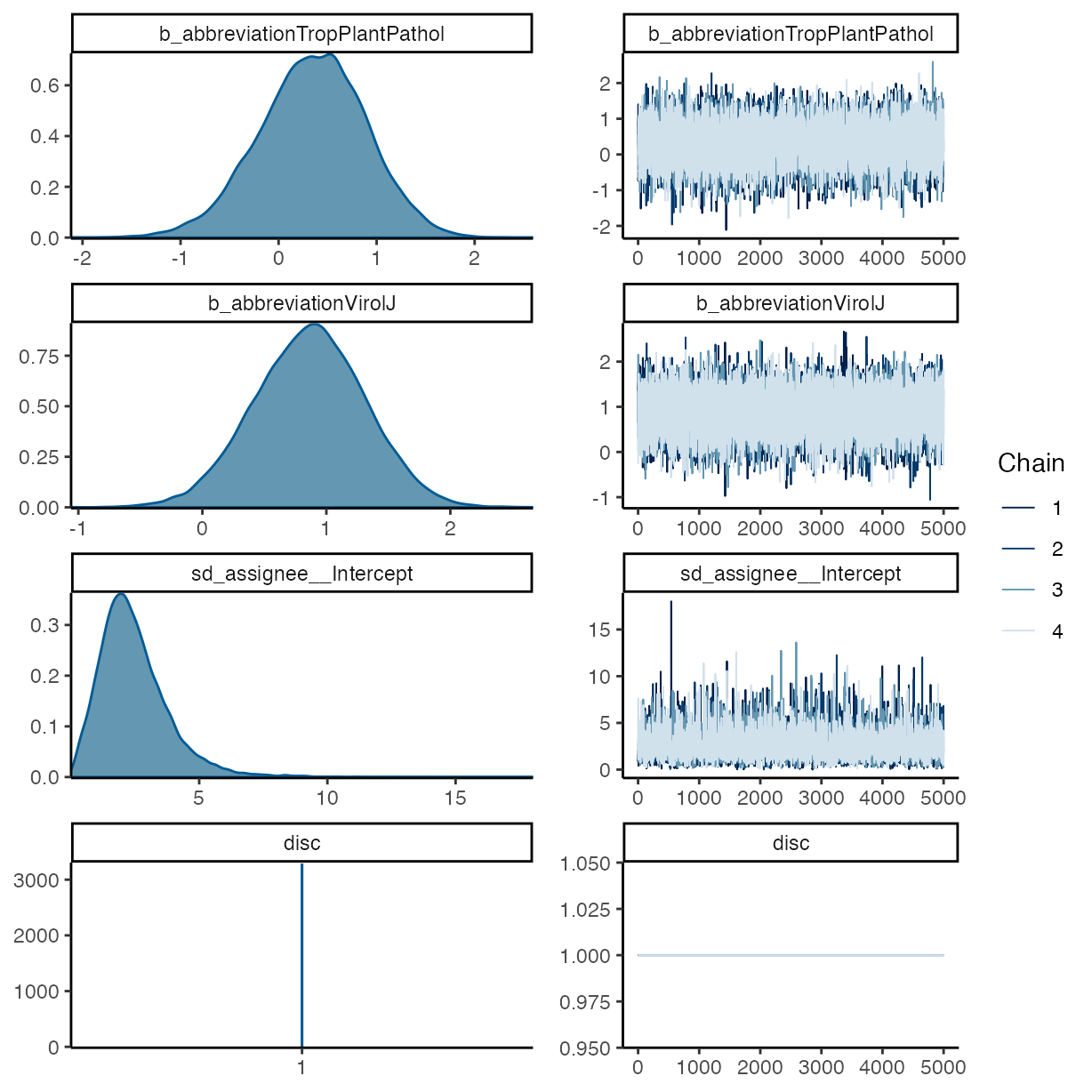
pp_check(m_f2, ndraws = 50, type = "bars")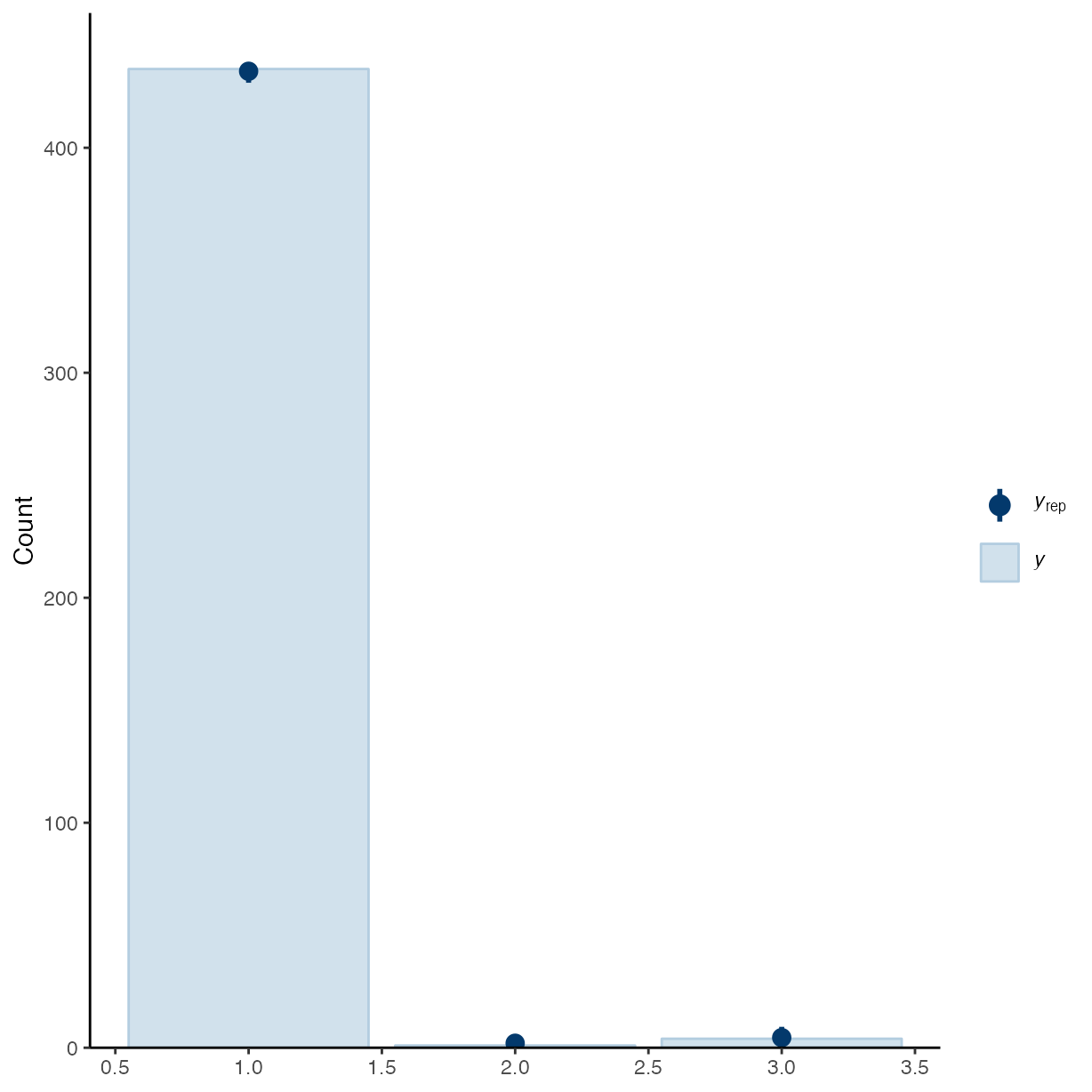
plot(equivalence_test(m_f2), range = c(-0.1, 0.1))
#> Picking joint bandwidth of 0.0611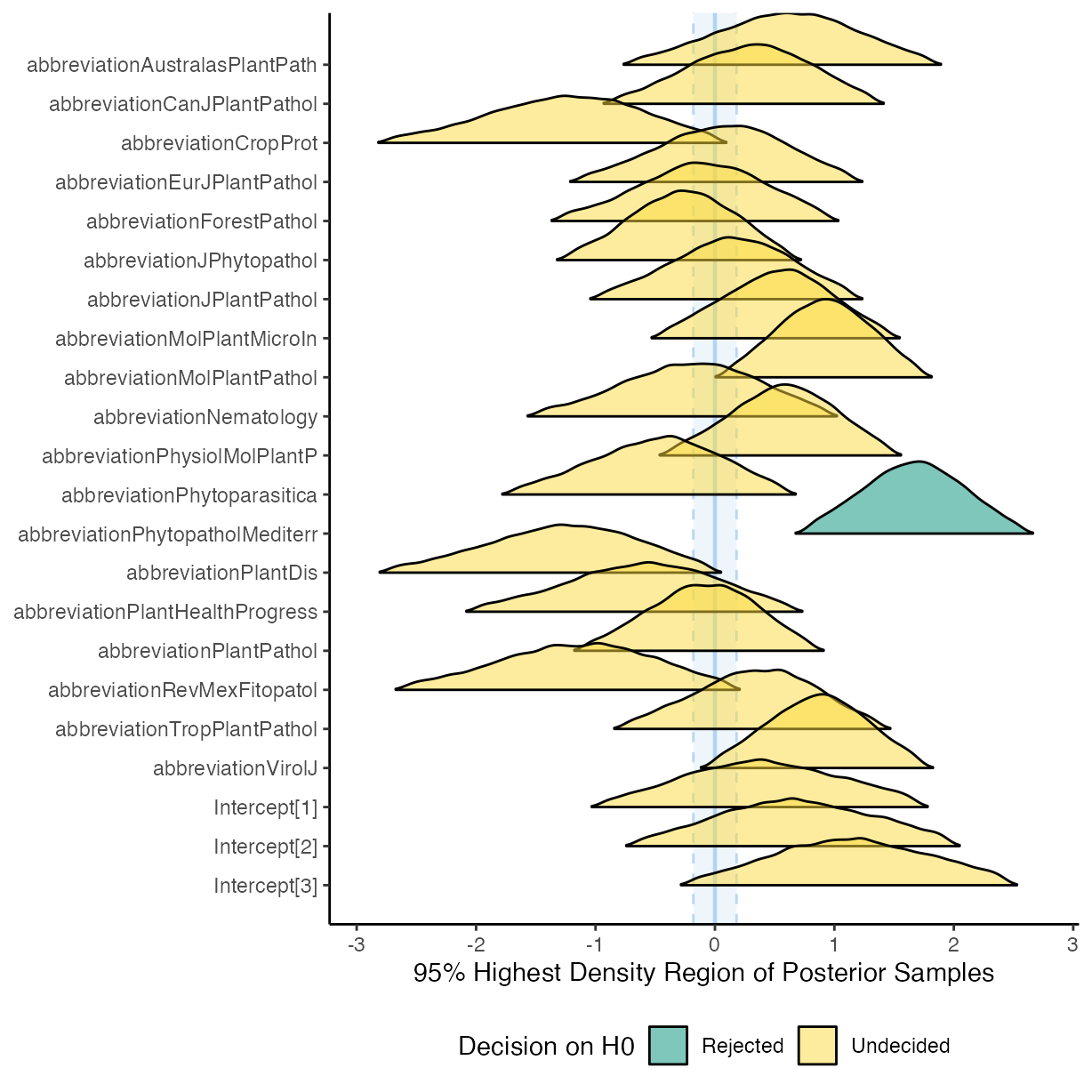
# pander(m_f2_report <- report(m_f2))
# m_f2_es <- report_effectsize(m_f2)Save Model Objects
Save the model objects for figures in the paper.
save(m_f1, file = here("inst/extdata/m_f1.Rda"))
save(m_f2, file = here("inst/extdata/m_f2.Rda"))
save(m_f1_report, file = here("inst/extdata/m_f1_report.Rda"))
save(m_f2_report, file = here("inst/extdata/m_f2_report.Rda"))
save(m_f1_es, file = here("inst/extdata/m_f1_es.Rda"))
save(m_f2_es, file = here("inst/extdata/m_f2_es.Rda"))Colophon
sessioninfo::session_info()
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.4.1 (2024-06-14)
#> os macOS Sonoma 14.6
#> system aarch64, darwin20
#> ui X11
#> language en
#> collate en_US.UTF-8
#> ctype en_US.UTF-8
#> tz Australia/Perth
#> date 2024-08-07
#> pandoc 3.3 @ /opt/homebrew/bin/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> abind 1.4-5 2016-07-21 [1] CRAN (R 4.4.0)
#> backports 1.5.0 2024-05-23 [1] CRAN (R 4.4.0)
#> bayesplot * 1.11.1 2024-02-15 [1] CRAN (R 4.4.0)
#> bayestestR * 0.14.0 2024-07-24 [1] CRAN (R 4.4.0)
#> bridgesampling 1.1-2 2021-04-16 [1] CRAN (R 4.4.0)
#> brms * 2.21.0 2024-03-20 [1] CRAN (R 4.4.1)
#> Brobdingnag 1.2-9 2022-10-19 [1] CRAN (R 4.4.0)
#> bslib 0.8.0 2024-07-29 [1] CRAN (R 4.4.0)
#> cachem 1.1.0 2024-05-16 [1] CRAN (R 4.4.0)
#> callr 3.7.6 2024-03-25 [1] CRAN (R 4.4.0)
#> cellranger 1.1.0 2016-07-27 [1] CRAN (R 4.4.0)
#> checkmate 2.3.2 2024-07-29 [1] CRAN (R 4.4.0)
#> cli 3.6.3 2024-06-21 [1] CRAN (R 4.4.0)
#> coda 0.19-4.1 2024-01-31 [1] CRAN (R 4.4.0)
#> codetools 0.2-20 2024-03-31 [2] CRAN (R 4.4.1)
#> colorspace 2.1-1 2024-07-26 [1] CRAN (R 4.4.0)
#> curl 5.2.1 2024-03-01 [1] CRAN (R 4.4.0)
#> datawizard 0.12.2 2024-07-21 [1] CRAN (R 4.4.0)
#> desc 1.4.3 2023-12-10 [1] CRAN (R 4.4.0)
#> digest 0.6.36 2024-06-23 [1] CRAN (R 4.4.0)
#> distributional 0.4.0 2024-02-07 [1] CRAN (R 4.4.0)
#> dplyr 1.1.4 2023-11-17 [1] CRAN (R 4.4.0)
#> emmeans 1.10.3 2024-07-01 [1] CRAN (R 4.4.0)
#> estimability 1.5.1 2024-05-12 [1] CRAN (R 4.4.0)
#> evaluate 0.24.0 2024-06-10 [1] CRAN (R 4.4.0)
#> fansi 1.0.6 2023-12-08 [1] CRAN (R 4.4.0)
#> farver 2.1.2 2024-05-13 [1] CRAN (R 4.4.0)
#> fastmap 1.2.0 2024-05-15 [1] CRAN (R 4.4.0)
#> fs 1.6.4 2024-04-25 [1] CRAN (R 4.4.0)
#> generics 0.1.3 2022-07-05 [1] CRAN (R 4.4.0)
#> ggplot2 * 3.5.1 2024-04-23 [1] CRAN (R 4.4.0)
#> ggridges 0.5.6 2024-01-23 [1] CRAN (R 4.4.0)
#> glue 1.7.0 2024-01-09 [1] CRAN (R 4.4.0)
#> gridExtra 2.3 2017-09-09 [1] CRAN (R 4.4.0)
#> gtable 0.3.5 2024-04-22 [1] CRAN (R 4.4.0)
#> here * 1.0.1 2020-12-13 [1] CRAN (R 4.4.0)
#> highr 0.11 2024-05-26 [1] CRAN (R 4.4.0)
#> htmltools 0.5.8.1 2024-04-04 [1] CRAN (R 4.4.0)
#> htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.4.0)
#> inline 0.3.19 2021-05-31 [1] CRAN (R 4.4.0)
#> insight 0.20.2 2024-07-13 [1] CRAN (R 4.4.0)
#> jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.4.0)
#> jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.4.0)
#> knitr 1.48 2024-07-07 [1] CRAN (R 4.4.0)
#> labeling 0.4.3 2023-08-29 [1] CRAN (R 4.4.0)
#> lattice 0.22-6 2024-03-20 [2] CRAN (R 4.4.1)
#> lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.4.0)
#> loo 2.8.0 2024-07-03 [1] CRAN (R 4.4.0)
#> magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.4.0)
#> MASS 7.3-60.2 2024-04-26 [2] CRAN (R 4.4.1)
#> Matrix 1.7-0 2024-04-26 [2] CRAN (R 4.4.1)
#> matrixStats 1.3.0 2024-04-11 [1] CRAN (R 4.4.0)
#> minty 0.0.1 2024-05-22 [1] CRAN (R 4.4.0)
#> multcomp 1.4-26 2024-07-18 [1] CRAN (R 4.4.0)
#> munsell 0.5.1 2024-04-01 [1] CRAN (R 4.4.0)
#> mvtnorm 1.2-5 2024-05-21 [1] CRAN (R 4.4.0)
#> nlme 3.1-164 2023-11-27 [2] CRAN (R 4.4.1)
#> pander * 0.6.5 2022-03-18 [1] CRAN (R 4.4.0)
#> pillar 1.9.0 2023-03-22 [1] CRAN (R 4.4.0)
#> pkgbuild 1.4.4 2024-03-17 [1] CRAN (R 4.4.0)
#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.4.0)
#> pkgdown 2.1.0 2024-07-06 [1] CRAN (R 4.4.0)
#> plyr 1.8.9 2023-10-02 [1] CRAN (R 4.4.0)
#> posterior 1.6.0 2024-07-03 [1] CRAN (R 4.4.0)
#> processx 3.8.4 2024-03-16 [1] CRAN (R 4.4.0)
#> ps 1.7.7 2024-07-02 [1] CRAN (R 4.4.0)
#> purrr 1.0.2 2023-08-10 [1] CRAN (R 4.4.0)
#> QuickJSR 1.3.1 2024-07-14 [1] CRAN (R 4.4.0)
#> R6 2.5.1 2021-08-19 [1] CRAN (R 4.4.0)
#> ragg 1.3.2 2024-05-15 [1] CRAN (R 4.4.0)
#> Rcpp * 1.0.13 2024-07-17 [1] CRAN (R 4.4.0)
#> RcppParallel 5.1.8 2024-07-06 [1] CRAN (R 4.4.0)
#> readODS 2.3.0 2024-05-26 [1] CRAN (R 4.4.0)
#> report * 0.4.0 2021-09-30 [1] CRAN (R 4.4.1)
#> Reproducibility.in.Plant.Pathology * 1.0.2 2024-08-06 [1] Github (openplantpathology/Reproducibility_in_Plant_Pathology@240170e)
#> reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.4.0)
#> rlang 1.1.4 2024-06-04 [1] CRAN (R 4.4.0)
#> rmarkdown 2.27 2024-05-17 [1] CRAN (R 4.4.0)
#> rprojroot 2.0.4 2023-11-05 [1] CRAN (R 4.4.0)
#> rstan 2.32.6 2024-03-05 [1] CRAN (R 4.4.1)
#> rstantools 2.4.0 2024-01-31 [1] CRAN (R 4.4.0)
#> rstudioapi 0.16.0 2024-03-24 [1] CRAN (R 4.4.0)
#> sandwich 3.1-0 2023-12-11 [1] CRAN (R 4.4.0)
#> sass 0.4.9 2024-03-15 [1] CRAN (R 4.4.0)
#> scales 1.3.0 2023-11-28 [1] CRAN (R 4.4.0)
#> see 0.8.5 2024-07-17 [1] CRAN (R 4.4.0)
#> sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.4.0)
#> StanHeaders 2.32.10 2024-07-15 [1] CRAN (R 4.4.0)
#> stringi 1.8.4 2024-05-06 [1] CRAN (R 4.4.0)
#> stringr 1.5.1 2023-11-14 [1] CRAN (R 4.4.0)
#> survival 3.6-4 2024-04-24 [2] CRAN (R 4.4.1)
#> systemfonts 1.1.0 2024-05-15 [1] CRAN (R 4.4.0)
#> tensorA 0.36.2.1 2023-12-13 [1] CRAN (R 4.4.0)
#> textshaping 0.4.0 2024-05-24 [1] CRAN (R 4.4.0)
#> TH.data 1.1-2 2023-04-17 [1] CRAN (R 4.4.0)
#> tibble 3.2.1 2023-03-20 [1] CRAN (R 4.4.0)
#> tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.4.0)
#> tidyselect 1.2.1 2024-03-11 [1] CRAN (R 4.4.0)
#> tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.4.0)
#> utf8 1.2.4 2023-10-22 [1] CRAN (R 4.4.0)
#> V8 4.4.2 2024-02-15 [1] CRAN (R 4.4.0)
#> vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.4.0)
#> withr 3.0.1 2024-07-31 [1] CRAN (R 4.4.0)
#> xfun 0.46 2024-07-18 [1] CRAN (R 4.4.0)
#> xtable 1.8-4 2019-04-21 [1] CRAN (R 4.4.0)
#> yaml 2.3.10 2024-07-26 [1] CRAN (R 4.4.0)
#> zip 2.3.1 2024-01-27 [1] CRAN (R 4.4.0)
#> zoo 1.8-12 2023-04-13 [1] CRAN (R 4.4.0)
#>
#> [1] /Users/283204f/Library/R/arm64/4.4/library
#> [2] /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/library
#>
#> ──────────────────────────────────────────────────────────────────────────────