Data Visualisation
Adam H. Sparks
2024-08-07
Source:vignettes/d_visualisation.Rmd
d_visualisation.RmdThis vignette documents a simple visualisation and tabulation of the data gathered from surveying 21 journals and 450 articles in the field of plant pathology for their openness and reproducibility.
Set-up Workspace
Load libraries used and setting the ggplot2 theme for the document.
library("tidyverse")
#> ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
#> ✔ dplyr 1.1.4 ✔ readr 2.1.5
#> ✔ forcats 1.0.0 ✔ stringr 1.5.1
#> ✔ ggplot2 3.5.1 ✔ tibble 3.2.1
#> ✔ lubridate 1.9.3 ✔ tidyr 1.3.1
#> ✔ purrr 1.0.2
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
#> ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
library("janitor")
#>
#> Attaching package: 'janitor'
#>
#> The following objects are masked from 'package:stats':
#>
#> chisq.test, fisher.test
library("pander")
library("Reproducibility.in.Plant.Pathology")
theme_set(theme_classic())Import the data and calculate the reproducibility score
rrpp <- import_notes()Individual Rating Scores
rrpp %>%
mutate(
comp_mthds_avail = as.numeric(as.character(comp_mthds_avail)),
data_avail = as.numeric(as.character(data_avail))
) %>%
pivot_longer(cols = c("comp_mthds_avail",
"data_avail")) %>%
ggplot(aes(x = as.factor(value))) +
geom_bar() +
ylab("Count") +
xlab("Criteria Score") +
ggtitle("Criteria scores by evaluator") +
facet_grid(name ~ assignee)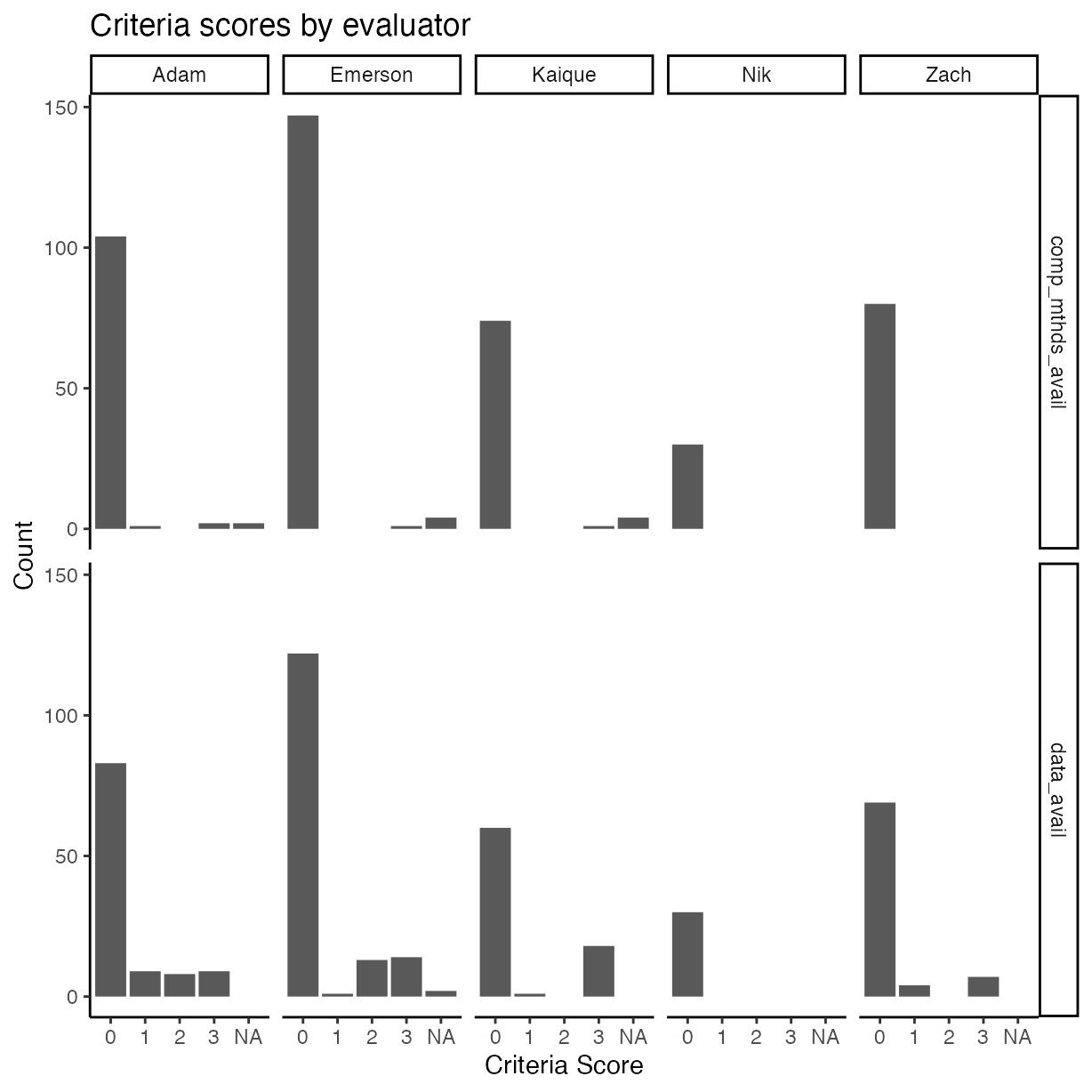
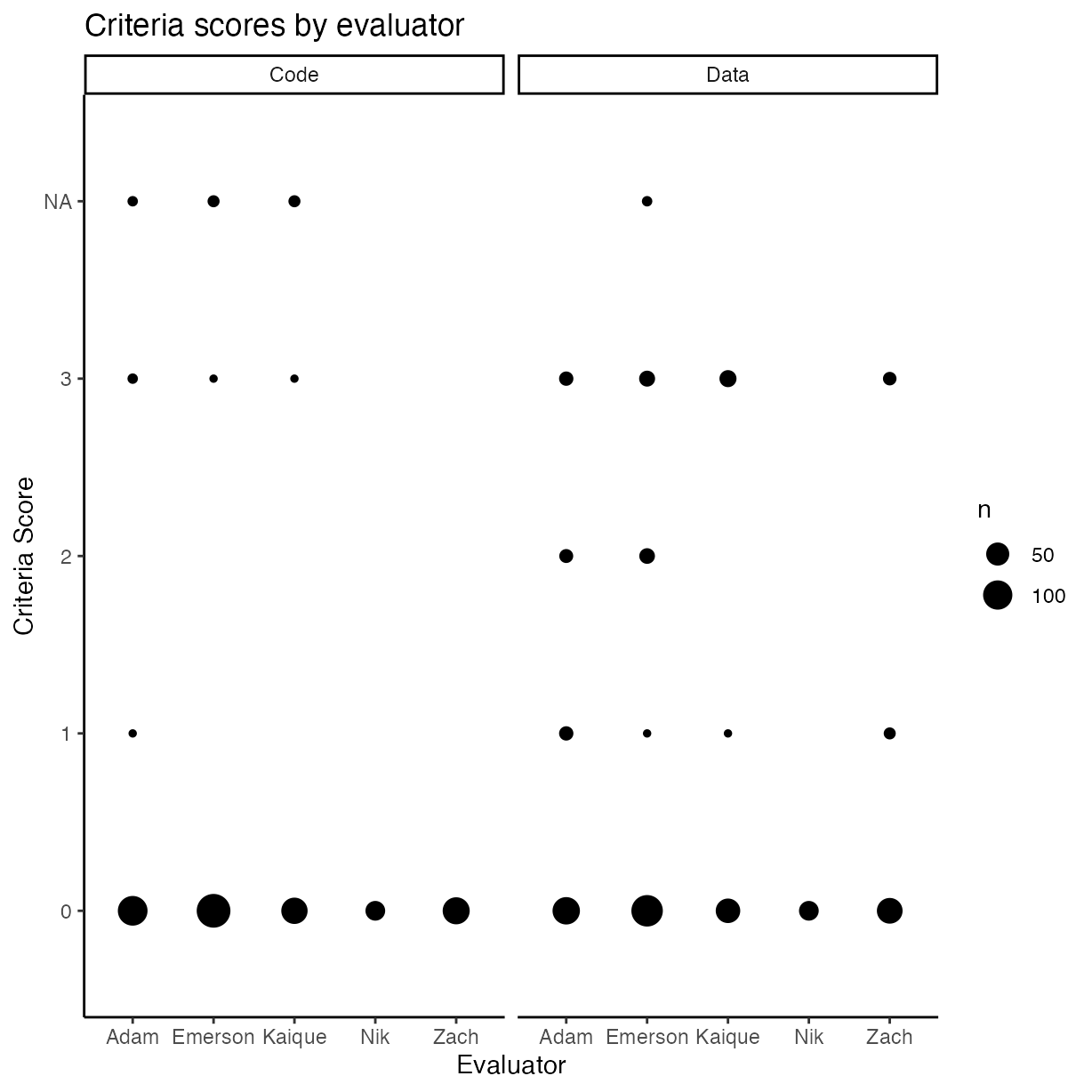
rrpp %>%
mutate(
comp_mthds_avail = as.numeric(as.character(comp_mthds_avail)),
data_avail = as.numeric(as.character(data_avail))
) %>%
pivot_longer(cols = c("comp_mthds_avail",
"data_avail")) %>%
ggplot(aes(y = as.factor(value), x = assignee)) +
geom_count() +
ylab("Criteria Score") +
xlab("Evaluator") +
ggtitle("Criteria scores by evaluator") +
facet_wrap(name ~ .,
labeller = labeller(name =
c(
"comp_mthds_avail" = "Code",
"data_avail" = "Data"
)))
Visualise evaluations
Computational methods available
Were the computational methods, scripts, source code, etc., made available?
ggplot(rrpp, aes(y = as.factor(comp_mthds_avail))) +
geom_bar() +
ggtitle("Computational Methods Availability") +
ylab("Score") +
labs(caption = "NA means that no computational methods were used to generate the data,\ne.g. only PCR results reported with no statistical analysis")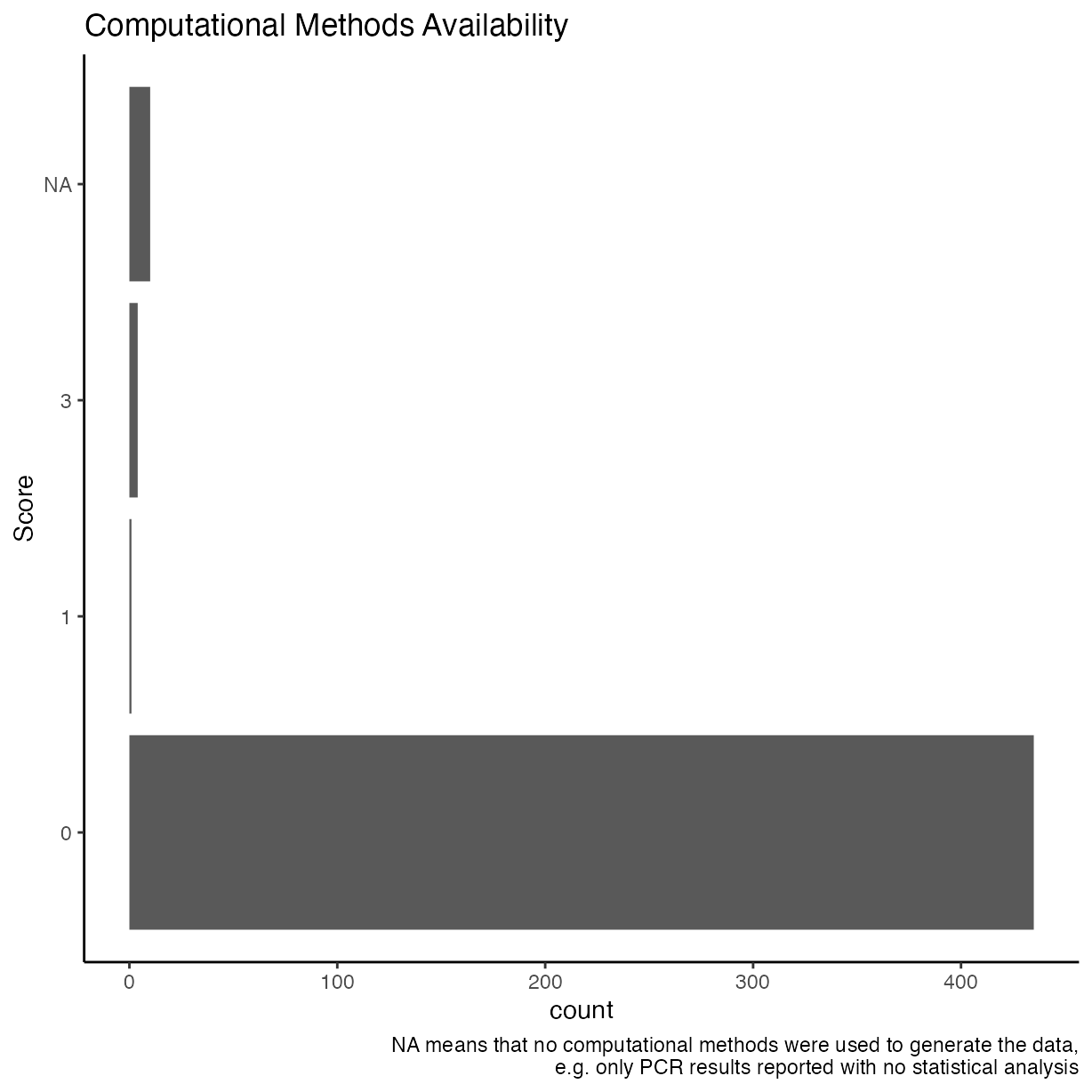
Software used (cited)
Count and sort top 10 software packages cited. There are likely others that are used, but they have not been properly cited or listed by the authors.
First create a tidy data frame of the data by unnesting software used as in many cases multiple software packages were used, so will end up with multiple rows for same article, one for each software.
rrpp_software <-
rrpp %>%
transform(software_used = strsplit(software_used, ",")) %>%
unnest(software_used) %>%
mutate(software_used = trimws(software_used)) %>%
mutate(software_used = toupper(software_used)) # convert all to uppercase to standardiseNow graph the top ten software packages used.
tab <- table(rrpp_software$software_used)
tab_s <- as.data.frame(sort(tab))
tab_s <-
tab_s %>%
arrange(desc(Freq)) %>%
filter(Freq %in% head(unique(Freq), 10)) %>%
rename("Software" = "Var1", "Frequency" = "Freq")
ggplot(tab_s, aes(y = Software, x = Frequency)) +
geom_bar(stat = "identity") +
ggtitle("Top 10 Software Used") +
xlab("Count") +
ylab("Software")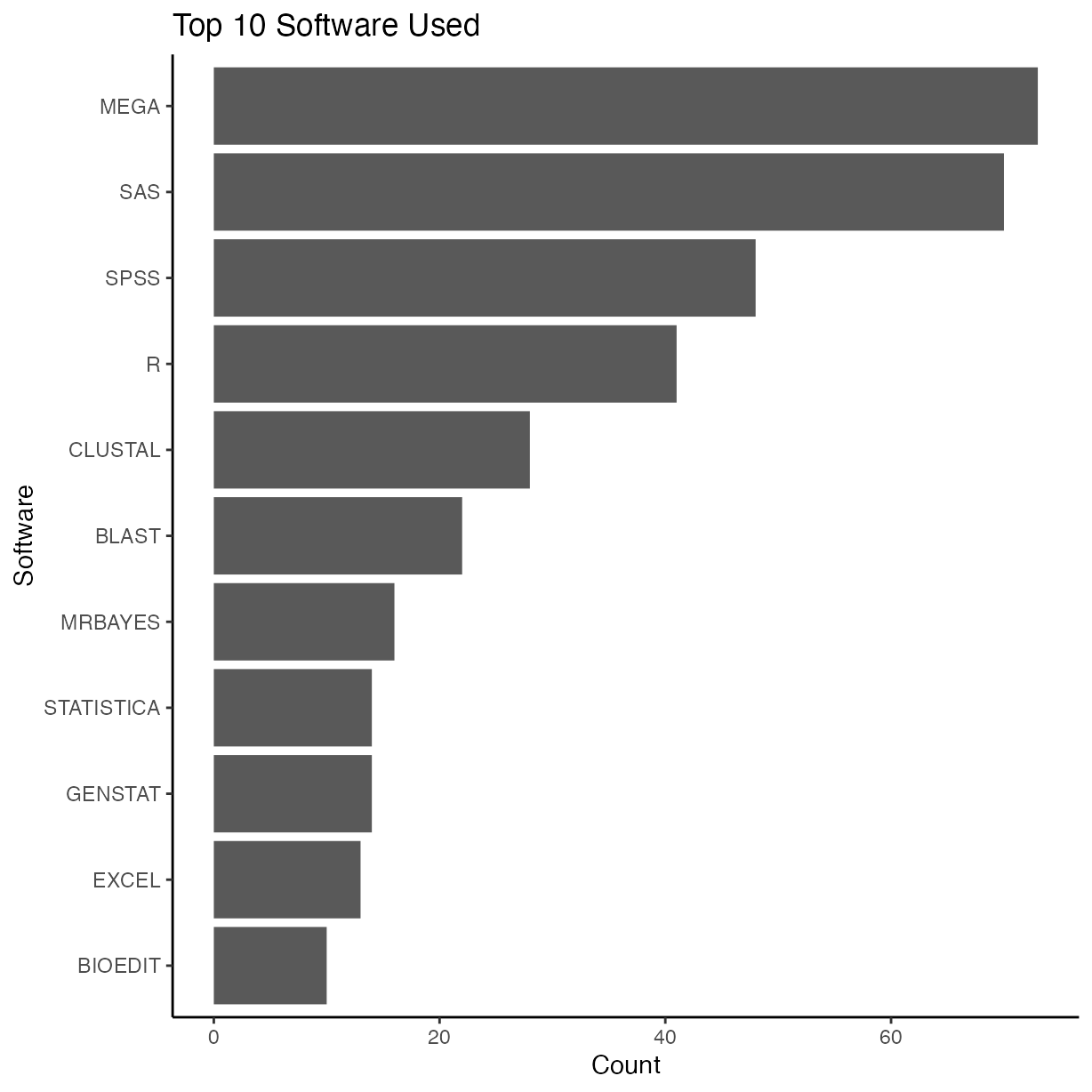
Data availability
Were the data made readily available?
ggplot(rrpp, aes(y = as.factor(data_avail))) +
geom_bar() +
ggtitle("Data Availability") +
ylab("Score")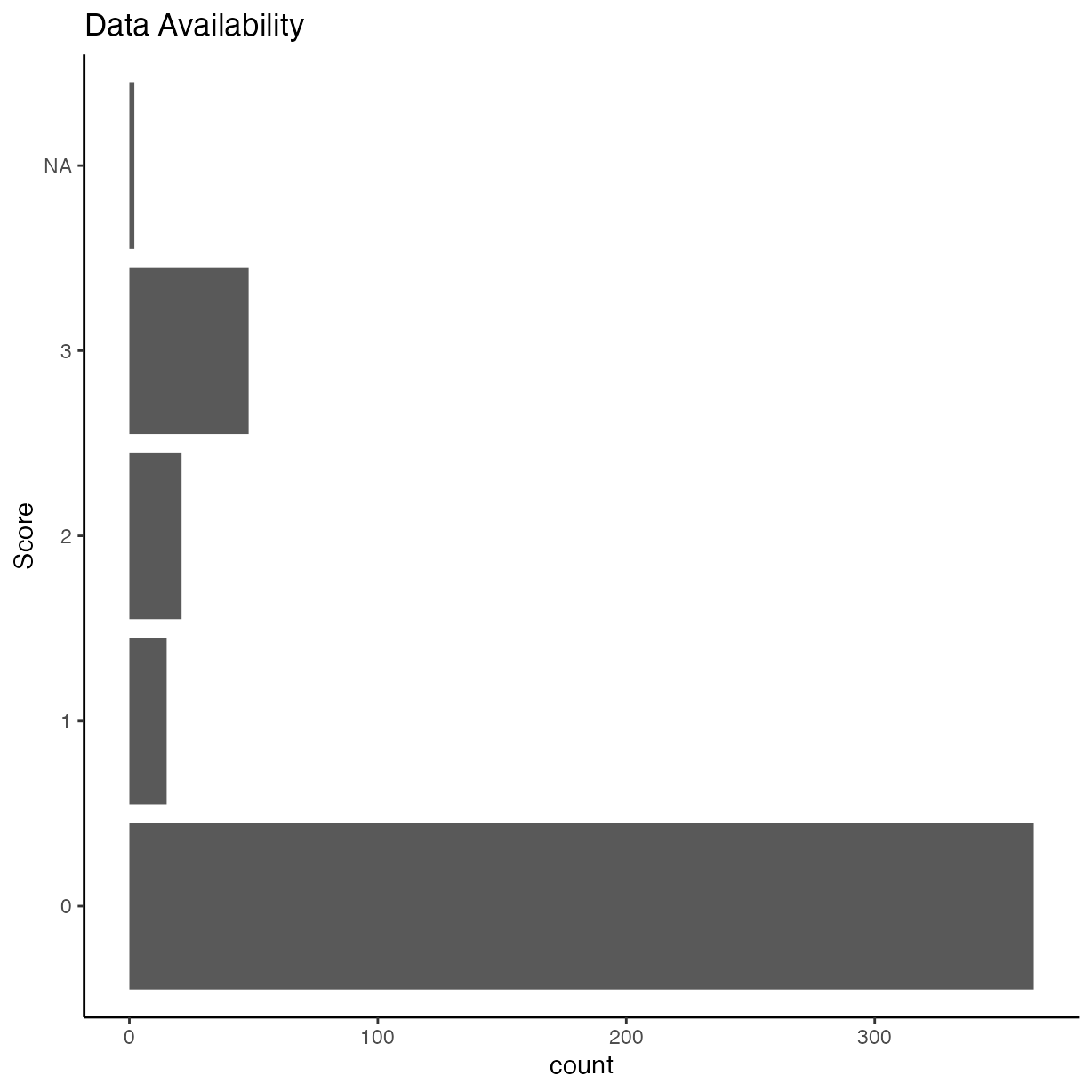
Scores by year
rrpp %>%
pivot_longer(cols = c("comp_mthds_avail", "data_avail")) %>%
ggplot(aes(x = year,
y = value)) +
geom_count() +
ylab("Score") +
xlab("Year") +
facet_grid(name ~ .)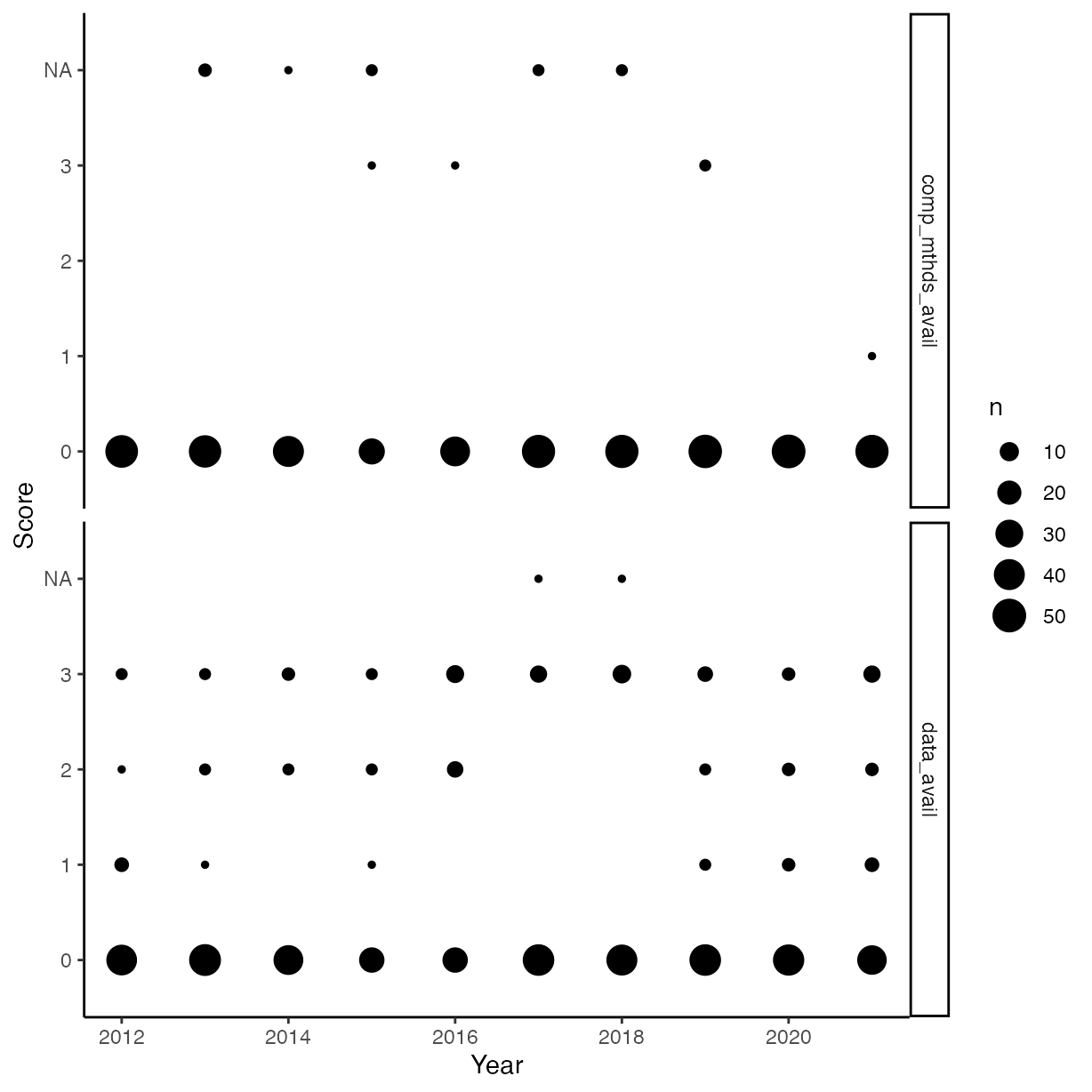
Visualise Probabilities
Code Availability
rrpp %>%
group_by(comp_mthds_avail) %>%
drop_na(comp_mthds_avail) %>%
count() %>%
mutate(pr_k = n / nrow(drop_na(rrpp, comp_mthds_avail))) %>%
ungroup() %>%
mutate(cum_pr_k = cumsum(pr_k)) %>%
ggplot(aes(x = as.numeric(as.character(comp_mthds_avail)), y = cum_pr_k,
fill = as.numeric(as.character(comp_mthds_avail)))) +
geom_line() +
geom_point(shape = 21,
colour = "grey92",
size = 2.5,
stroke = 1) +
scale_y_continuous("cumulative proportion", breaks = c(0, .5, 1)) +
scale_fill_gradient() +
coord_cartesian(ylim = c(0, 1)) +
theme(legend.position = "none") +
xlab("Computational Methods Availability Score")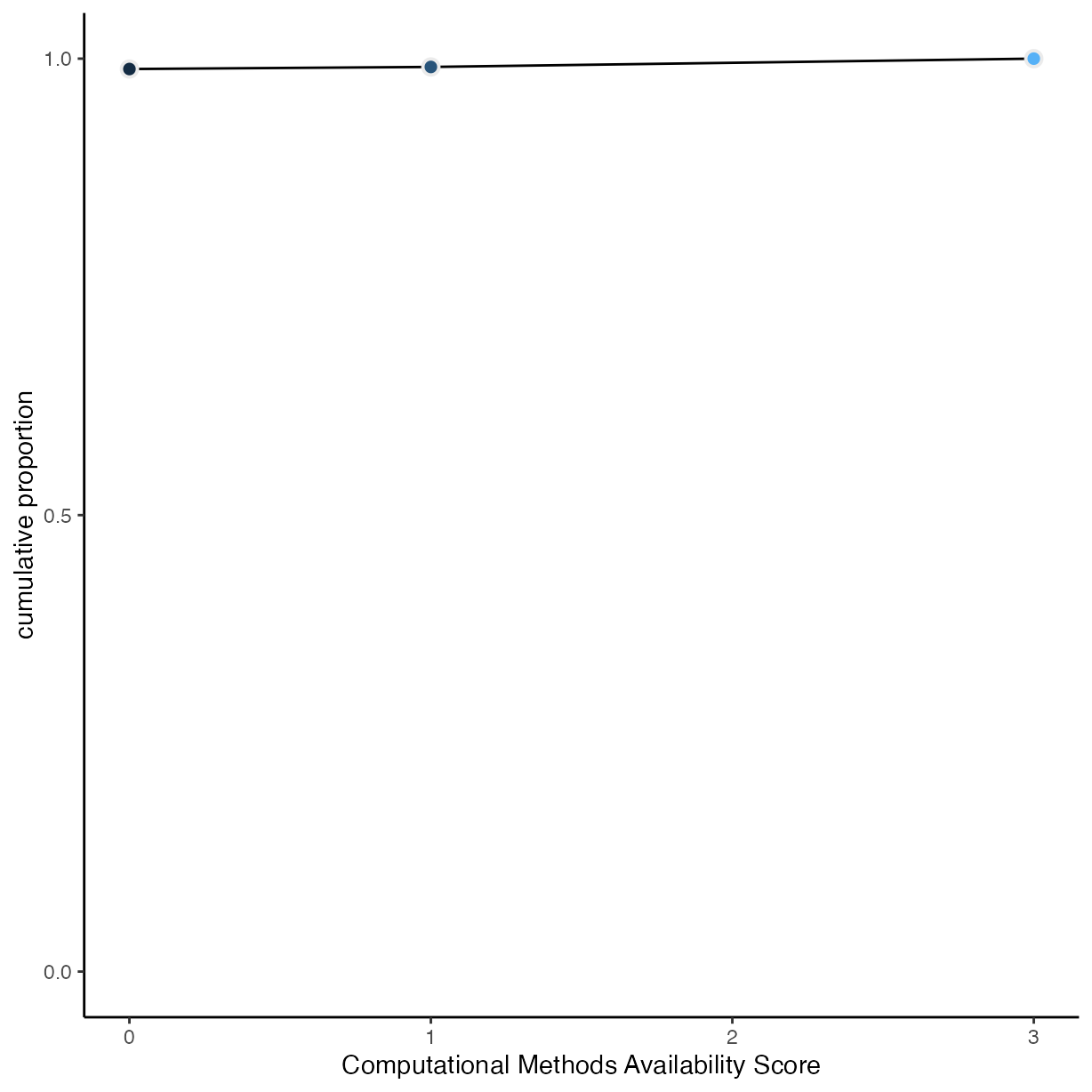
# McElreath's convenience function from page 335
logit <- function(x) log(x / (1 - x))
rrpp %>%
group_by(comp_mthds_avail) %>%
drop_na(comp_mthds_avail) %>%
count() %>%
mutate(pr_k = n / nrow(drop_na(rrpp, comp_mthds_avail))) %>%
ungroup() %>%
mutate(cum_pr_k = cumsum(pr_k)) %>%
filter(comp_mthds_avail < 3) %>%
ggplot(aes(
x = as.numeric(as.character(comp_mthds_avail)),
y = logit(cum_pr_k),
fill = as.numeric(as.character(comp_mthds_avail))
)) +
geom_line() +
geom_point(
shape = 21,
colour = "grey92",
size = 2.5,
stroke = 1
) +
coord_cartesian(xlim = c(0, 3)) +
ylab("log-cumulative-odds") +
xlab("Code Availability") +
scale_fill_gradient() +
theme(legend.position = "none")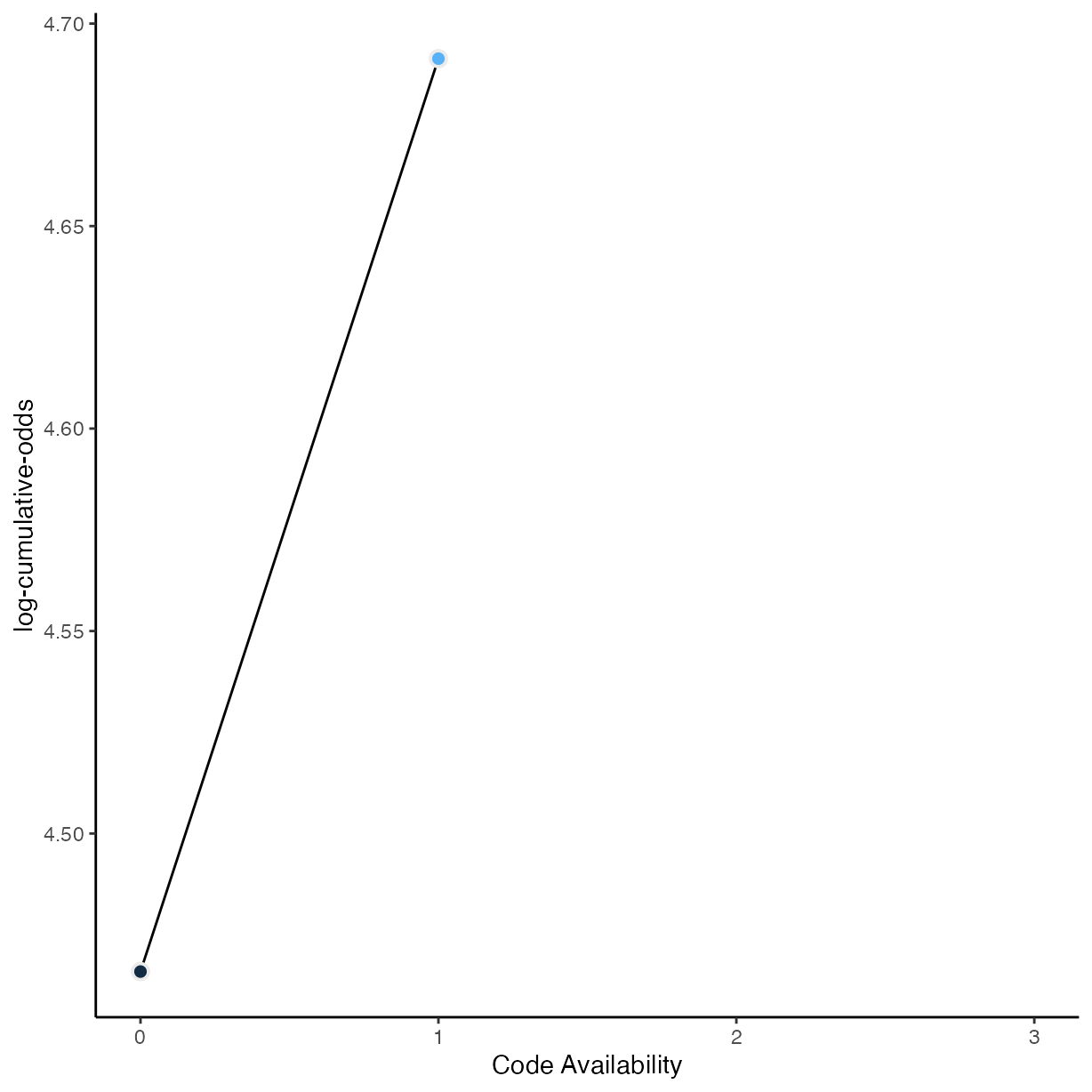
Data Availability
rrpp %>%
group_by(data_avail) %>%
drop_na(data_avail) %>%
count() %>%
mutate(pr_k = n / nrow(drop_na(rrpp, data_avail))) %>%
ungroup() %>%
mutate(cum_pr_k = cumsum(pr_k)) %>%
ggplot(aes(x = as.numeric(as.character(data_avail)), y = cum_pr_k,
fill = as.numeric(as.character(data_avail)))) +
geom_line() +
geom_point(shape = 21,
colour = "grey92",
size = 2.5,
stroke = 1) +
scale_y_continuous("cumulative proportion", breaks = c(0, .5, 1)) +
scale_fill_gradient() +
coord_cartesian(ylim = c(0, 1)) +
theme(legend.position = "none") +
xlab("Data Availability Score")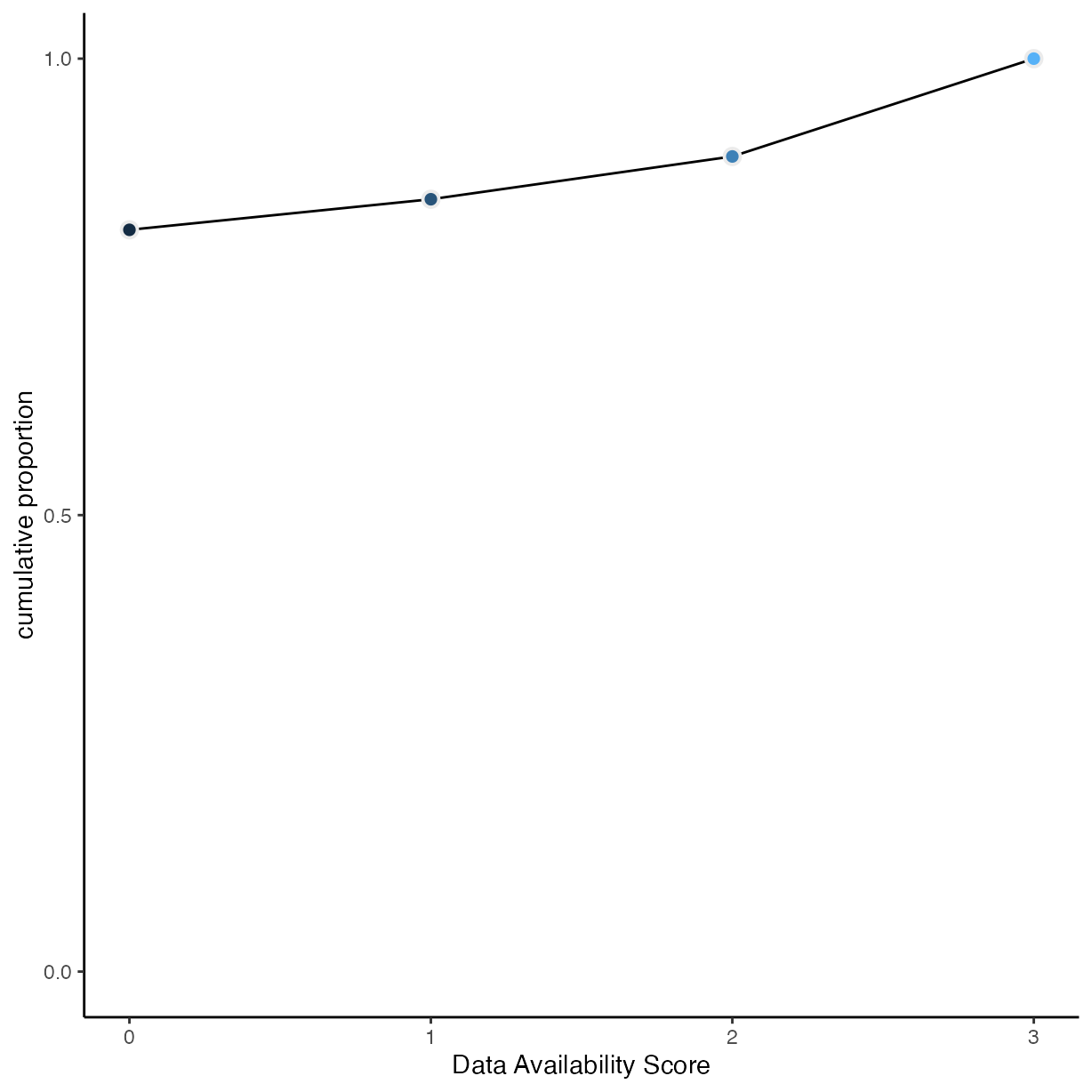
rrpp %>%
group_by(data_avail) %>%
drop_na(data_avail) %>%
count() %>%
mutate(pr_k = n / nrow(drop_na(rrpp, data_avail))) %>%
ungroup() %>%
mutate(cum_pr_k = cumsum(pr_k)) %>%
filter(data_avail < 3) %>%
ggplot(aes(
x = as.numeric(as.character(data_avail)),
y = logit(cum_pr_k),
fill = as.numeric(as.character(data_avail))
)) +
geom_line() +
geom_point(
shape = 21,
colour = "grey92",
size = 2.5,
stroke = 1
) +
coord_cartesian(xlim = c(0, 3)) +
ylab("log-cumulative-odds") +
xlab("Code Availability") +
scale_fill_gradient() +
theme(legend.position = "none")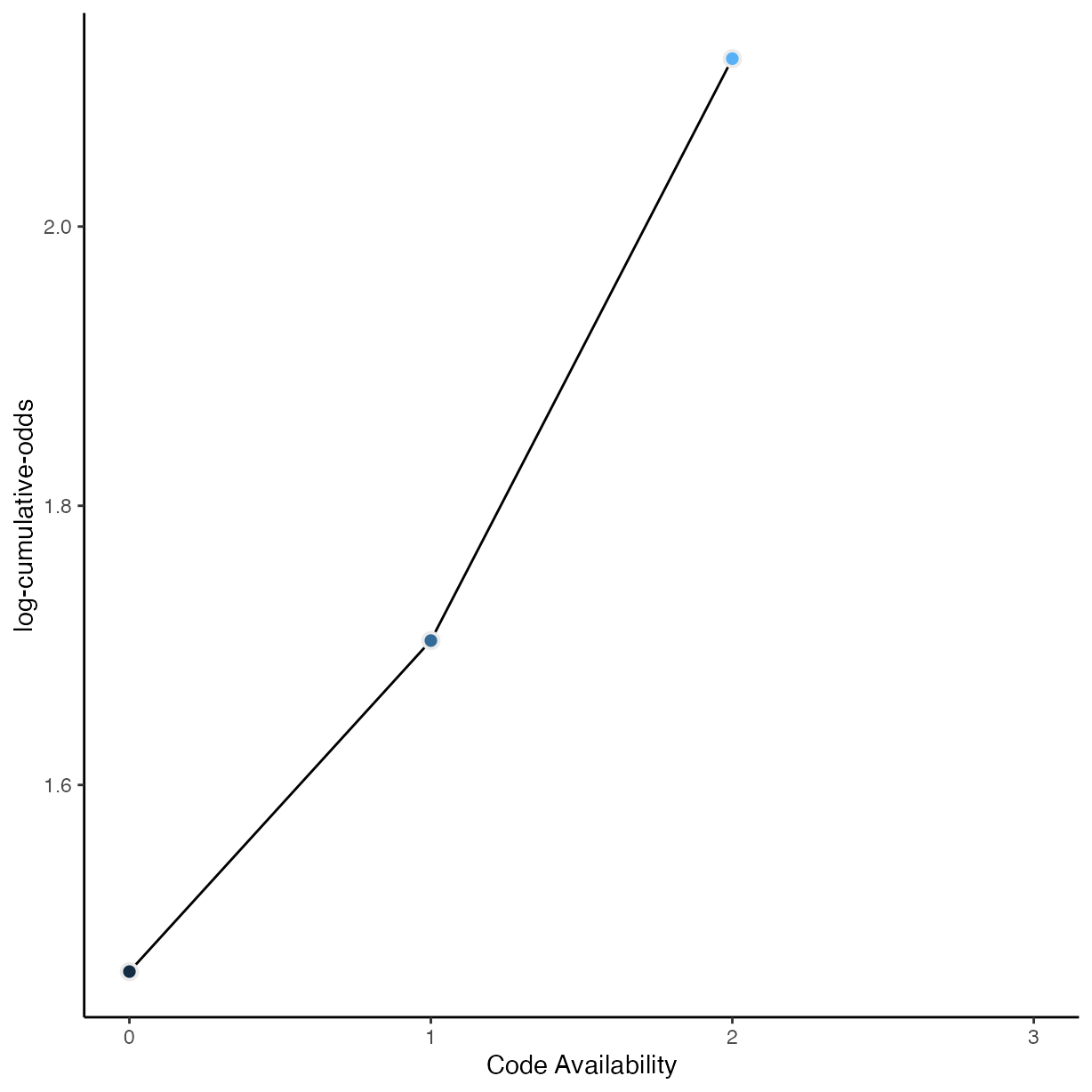
Tables
Table of journals surveyed
rrpp_journals <- tabyl(rrpp, journal)[, -3]
names(rrpp_journals) <- c("Journal", "n")
pander(rrpp_journals)| Journal | n |
|---|---|
| Australasian Plant Pathology | 11 |
| Canadian Journal of Plant Pathology | 19 |
| Crop Protection | 23 |
| European Journal of Plant Pathology | 19 |
| Forest Pathology | 18 |
| Journal of General Plant Pathology | 24 |
| Journal of Phytopathology | 19 |
| Journal of Plant Pathology | 23 |
| Molecular Plant Pathology | 29 |
| Molecular Plant-Microbe Interactions | 22 |
| Nematology | 16 |
| Physiological and Molecular Plant Pathology | 24 |
| Phytoparasitica | 23 |
| Phytopathologia Mediterranea | 20 |
| Phytopathology | 28 |
| Plant Disease | 24 |
| Plant Health Progress | 16 |
| Plant Pathology | 30 |
| Revista Mexicana de Fitopatología | 19 |
| Tropical Plant Pathology | 18 |
| Virology Journal (Plant Viruses Section) | 25 |