5 Powdery mildew severity meta-analysis
An additional question to our main aim: does spray schedule have a similar effect on disease severity?
This model will be similar to the grain yield meta-analysis and use the main variables:
-
Powdery mildew severity, the response variable (1-9 scale)
-
Trial, which resolves combinations of categorical random variables:
- year
- location
- row spacing
- cultivar
- year
spray_management, a moderator to evaluate the difference in effect size attributed to fungicide application timing.
id, random variable indicating each treatment is independent
-
Load libraries and data.
if (!require("pacman"))
install.packages("pacman")
pacman::p_load(tidyverse,
kableExtra,
RColorBrewer,
metafor,
here,
netmeta,
multcomp,
dplyr,
flextable,
gam,
clubSandwich)
source(here("R/reportP.R"))
# Data
PM_dat_D <-
read.csv("cache/PM_disease_clean_data.csv", stringsAsFactors = FALSE)
# functions
logit <- function(x){
log((x)/(1 - x))
}
inv_logit <- function(x){
exp(x)/(1 + exp(x))
}5.0.1 Define Trial
PM_dat_D <-
PM_dat_D %>%
mutate(trial = paste(trial_ref,
year,
location,
host_genotype,
row_spacing,
sep = "_")) 5.0.2 Inspect PM severity for normality
Given the meta-analysis assumes powdery mildew severity observations to be a univariate normal distribution \(Y_i = N(\mu , \sigma)\), with a mean \(\mu\) and standard deviation \(\sigma\). We need to inspect severity for a normality.
hist(PM_dat_D$PM_final_severity)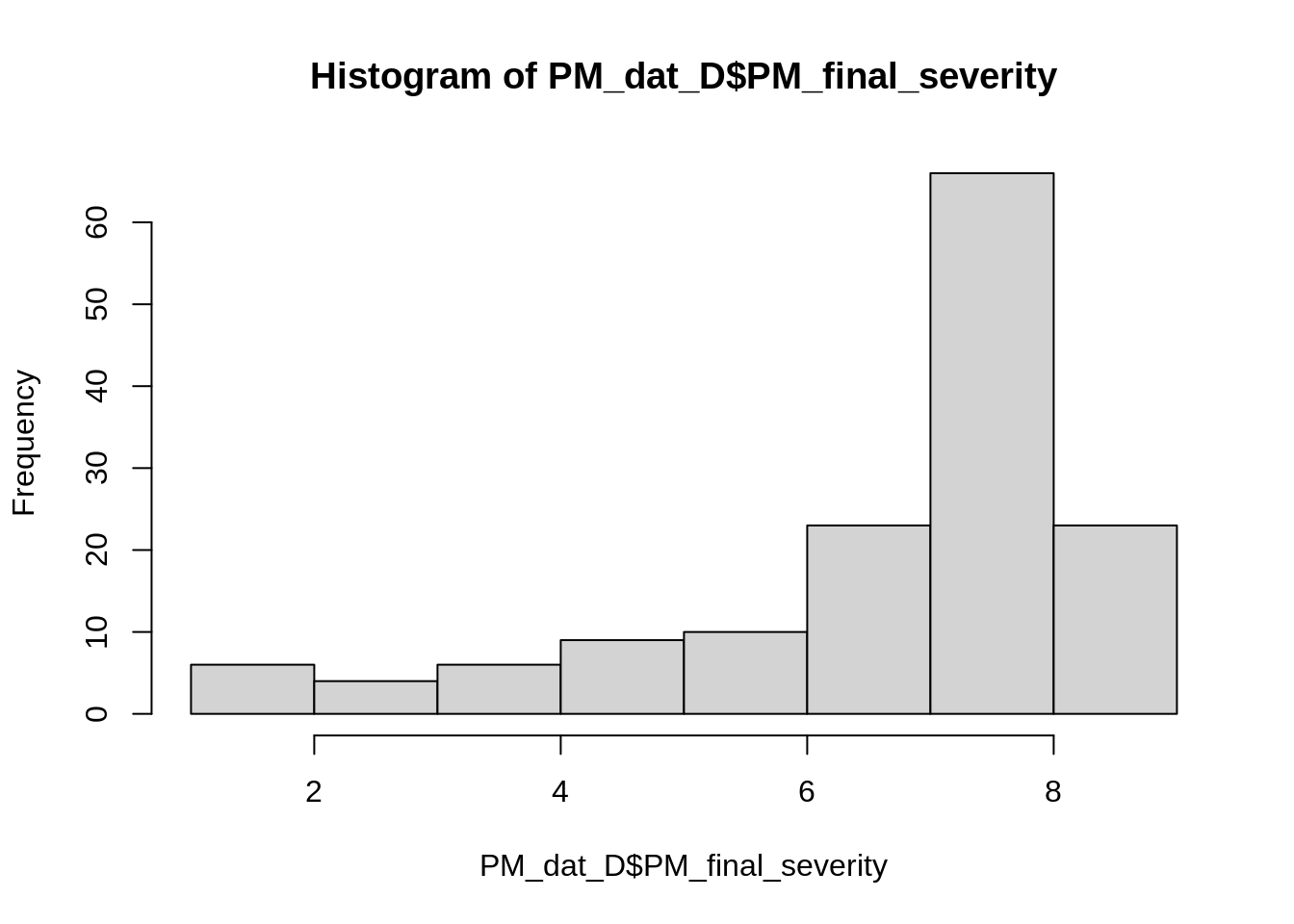
summary(PM_dat_D$PM_final_severity)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 1.000 6.367 7.667 6.933 8.000 9.000We can see here that powdery mildew severity observations are skewed towards the top of the scale, eight and nine in this case.
We can improve the normality by transforming the effect size, disease severity, by squaring.
PM_dat_D$yi_square <- PM_dat_D$PM_final_severity ^2
PM_dat_D$vi_square <- (PM_dat_D$vi +1)^2
# Note: Here I added 1 to the variance before it was squared. This is because squaring numbers below one makes the number smaller or remain zero. This does not change the meta-analysis mean estimates but does increase heterogeneity and tao._
hist(PM_dat_D$yi_square)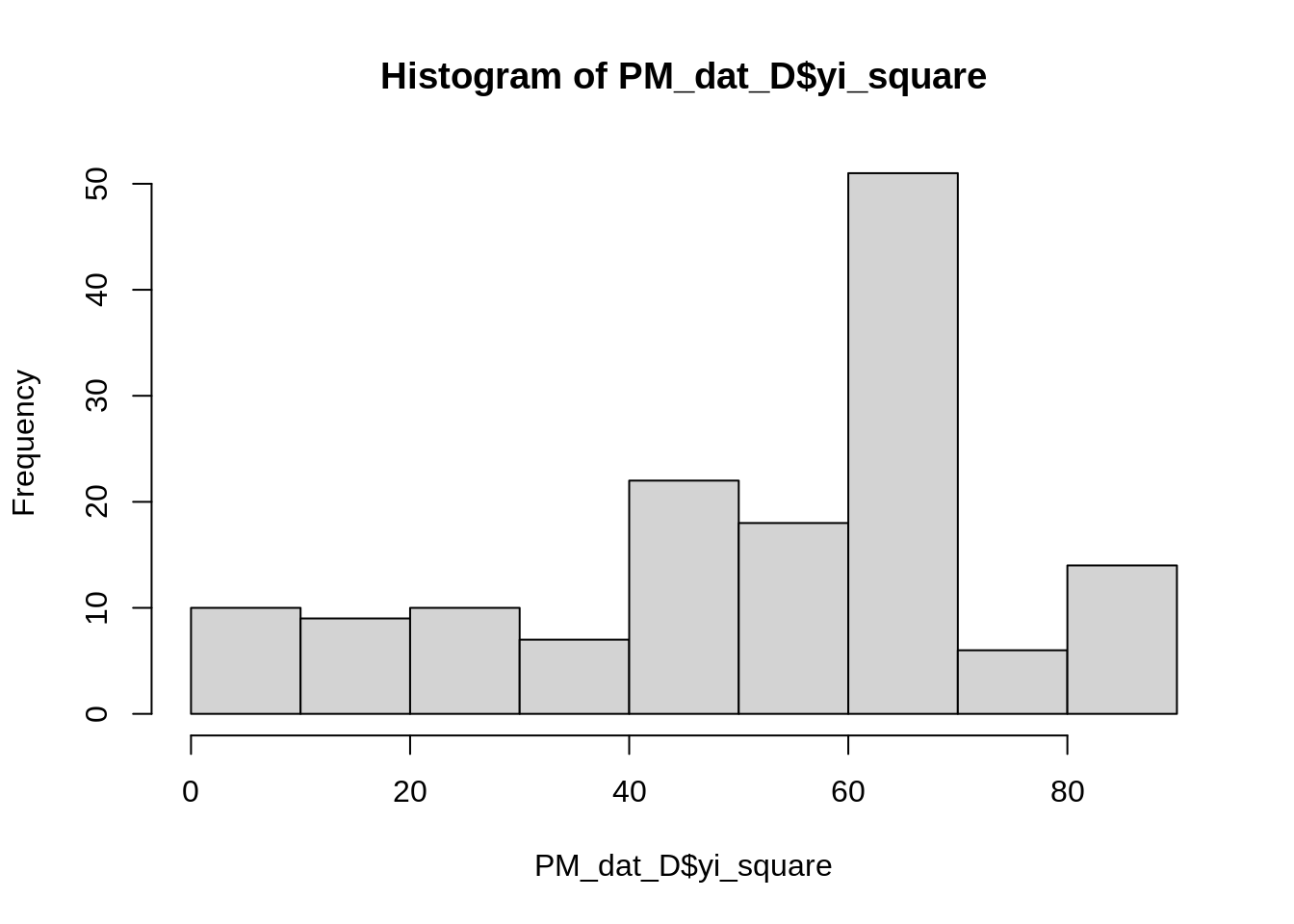
summary(PM_dat_D$yi_square)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 1.00 40.54 58.78 51.62 64.00 81.00Another transformation which might be more suitable for a discrete response is either a log() or logit() transformation.
However, first we should convert the scale from 1 - 9 to between 0 and 1.
5.0.3 Transform severity response
Let us also transform the severity onto a percentage scale so we can compare the results of meta-analyses with no transformation, squared severity, and percent severity.
This conversion from ordinal scale to a severity rating is described in Table 2 of the paper.
# Percent severity conversion
# define ordinal conversion from scale to proportion
s1 <- 0
s2 <- 0.33 * 0.5
s3 <- 0.5 * 0.87
s4 <- 0.66 * 0.75
s5 <- 0.66 * 0.87
s6 <- 0.66 * 1
s7 <- 0.75 * 1
s8 <- 1 * 0.87
s9 <- 1
x <- c(s1, s2, s3, s4, s5, s6, s7, s8, s9)
y <- c(1:9)
a = data.frame(x, y)
plot(a)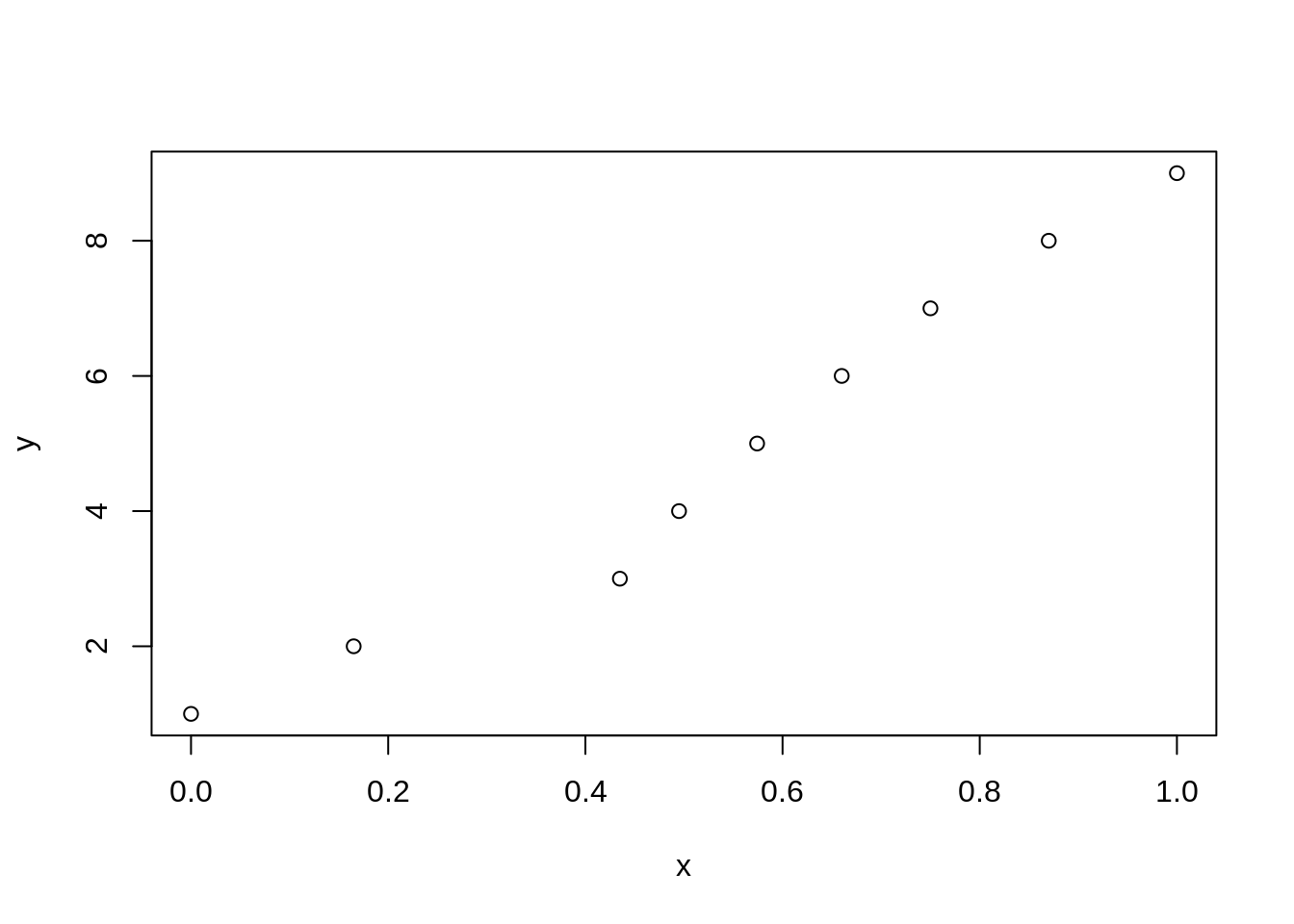
We can fit a GAM curve between the 1 - 9 scale and the new scale between 0 and 1.
# fit a curve using a gam with spline value 0.01
g1 <- gam(x ~ s(y, spar = 0.1), family = gaussian)
# View plot of gam
plot(x = predict(g1,
data.frame(y = seq(1, 9, by = 0.1))), y = seq(1, 9, by = 0.1))
points(x, y, col = "red", pch = 16)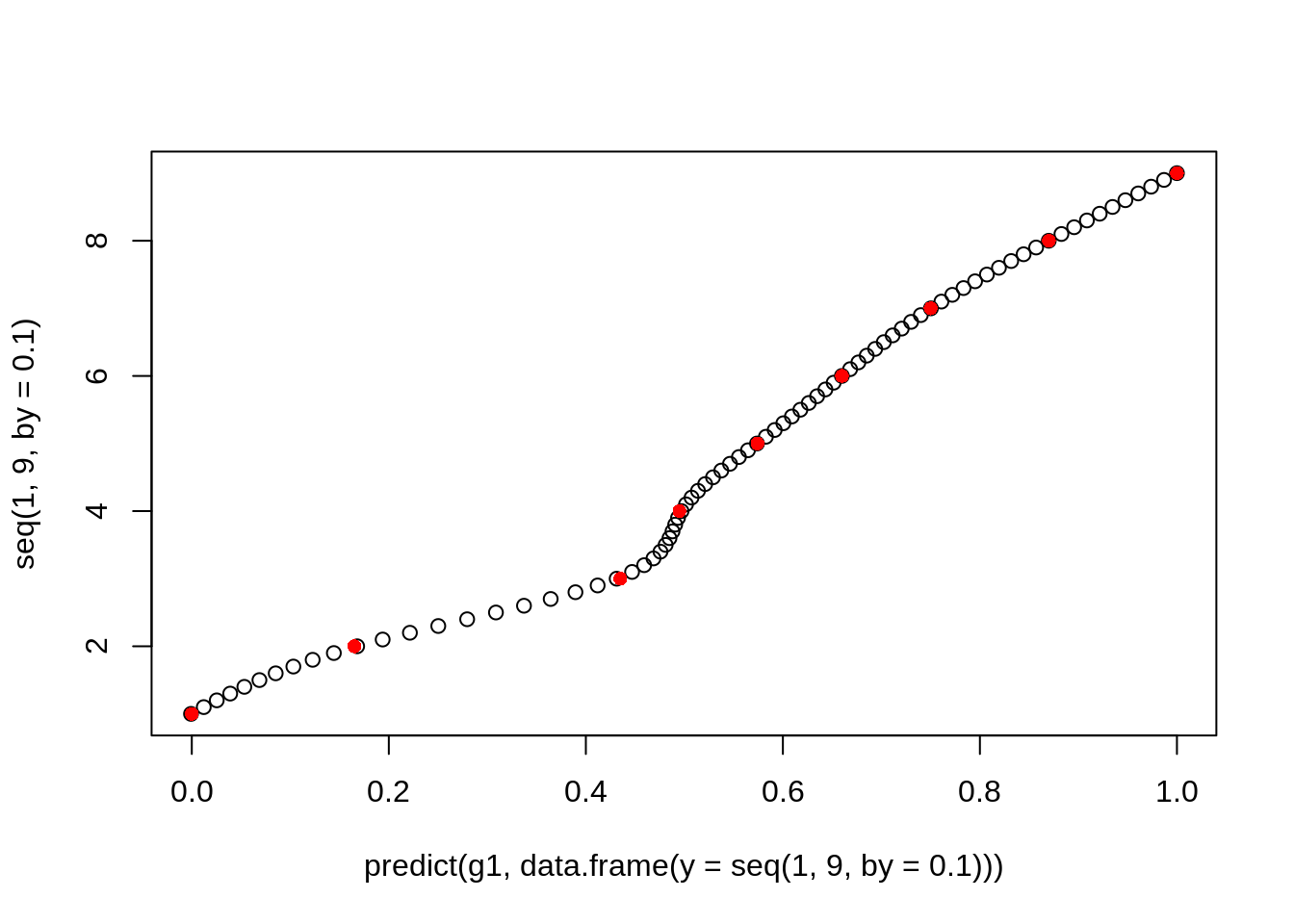
# predict values for dataset
new_sev <- predict(object = g1,
newdata = data.frame(y = PM_dat_D[, "PM_final_severity"]))
# Check fit
plot(x = new_sev, y = PM_dat_D[, "PM_final_severity"])
points(x, y, col = "red", pch = 16)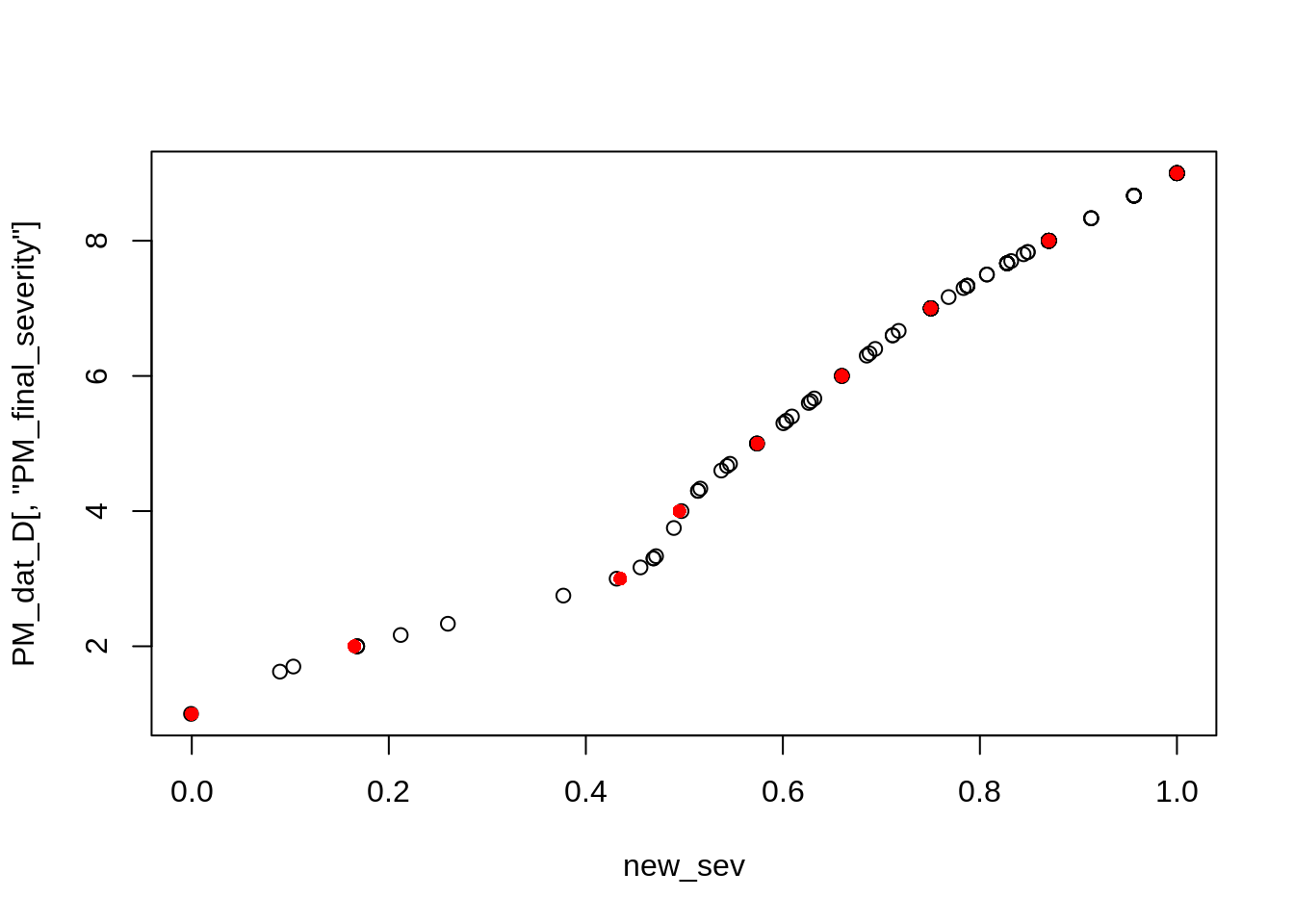
# Allocate new variable percent severity
PM_dat_D$yi_percent <- new_sev
# Conversion of variance
# Here I add the variance and mean severity and predict the conversion value
# then subtract the mean severity to obtain the difference as the sample variance
PM_dat_D$vi_percent <- predict(object = g1,
newdata = data.frame(y = PM_dat_D[, "vi"] + PM_dat_D[, "PM_final_severity"])) - PM_dat_D$yi_percent
# create log transformed values
PM_dat_D$yi_log <- log(PM_dat_D$yi_percent)## Warning in log(PM_dat_D$yi_percent): NaNs produced
PM_dat_D$vi_log <-
(PM_dat_D$vi_percent) / (PM_dat_D$replicates * PM_dat_D$yi_percent ^ 2)
# Correct Inf to 0
PM_dat_D[is.na(PM_dat_D$yi_log), "yi_log"] <- 0
PM_dat_D[PM_dat_D$vi_log == -Inf, "vi_log"] <- 0.001
#logit transformed percentages
PM_dat_D$yi_logit <- logit(PM_dat_D$yi_percent)## Warning in log((x)/(1 - x)): NaNs produced
# logit transformed variance
PM_dat_D$vi_logit <-
(PM_dat_D$vi_percent) /
(PM_dat_D$yi_percent * (1 - PM_dat_D$yi_percent)) ^ 2
# Correct -Inf to zeros
PM_dat_D[is.na(PM_dat_D$yi_logit), "yi_logit"] <- 05.1 Disease severity spray schedule meta-analysis
First let’s look at the results of the meta-analysis without transformation
PMsev_mv <- rma.mv(
yi = PM_final_severity,
vi,
mods = ~ spray_management,
method = "ML",
random = list(~ spray_management | trial, ~ 1 | id),
struct = "UN",
#control = list(optimizer = "optim"),
data = PM_dat_D
)## Warning: There are outcomes with non-positive sampling variances.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.
summary(PMsev_mv)##
## Multivariate Meta-Analysis Model (k = 147; method: ML)
##
## logLik Deviance AIC BIC AICc
## -145.7909 Inf 345.5818 426.3235 358.2877
##
## Variance Components:
##
## estim sqrt nlvls fixed factor
## sigma^2 0.0369 0.1922 147 no id
##
## outer factor: trial (nlvls = 22)
## inner factor: spray_management (nlvls = 6)
##
## estim sqrt k.lvl fixed level
## tau^2.1 1.5788 1.2565 37 no control
## tau^2.2 2.9285 1.7113 13 no Early
## tau^2.3 0.6798 0.8245 15 no Late
## tau^2.4 0.0619 0.2488 13 no Late_plus
## tau^2.5 4.2455 2.0605 28 no Recommended
## tau^2.6 6.9310 2.6327 41 no Recommended_plus
##
## rho.cntr rho.Erly rho.Late rho.Lt_p rho.Rcmm rho.Rcm_
## control 1 0.8287 0.9646 0.9977 0.9402 0.5966
## Early 0.8287 1 0.9341 0.0000 0.8077 0.9436
## Late 0.9646 0.9341 1 0.9720 0.8830 0.7691
## Late_plus 0.9977 0.0000 0.9720 1 0.9537 0.6407
## Recommended 0.9402 0.8077 0.8830 0.9537 1 0.6002
## Recommended_plus 0.5966 0.9436 0.7691 0.6407 0.6002 1
## cntr Erly Late Lt_p Rcmm Rcm_
## control - no no no no no
## Early 7 - no yes no no
## Late 9 6 - no no no
## Late_plus 4 0 1 - no no
## Recommended 18 6 9 1 - no
## Recommended_plus 16 6 9 1 16 -
##
## Test of Moderators (coefficients 2:6):
## QM(df = 5) = 163.0624, p-val < .0001
##
## Model Results:
##
## estimate se zval pval ci.lb
## intrcpt 7.6729 0.2766 27.7415 <.0001 7.1308
## spray_managementEarly -1.0439 0.2582 -4.0434 <.0001 -1.5500
## spray_managementLate -0.7455 0.1360 -5.4832 <.0001 -1.0120
## spray_managementLate_plus -1.3633 0.2383 -5.7207 <.0001 -1.8304
## spray_managementRecommended -1.1069 0.2595 -4.2659 <.0001 -1.6155
## spray_managementRecommended_plus -3.2476 0.5445 -5.9645 <.0001 -4.3148
## ci.ub
## intrcpt 8.2150 ***
## spray_managementEarly -0.5379 ***
## spray_managementLate -0.4790 ***
## spray_managementLate_plus -0.8962 ***
## spray_managementRecommended -0.5983 ***
## spray_managementRecommended_plus -2.1804 ***
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1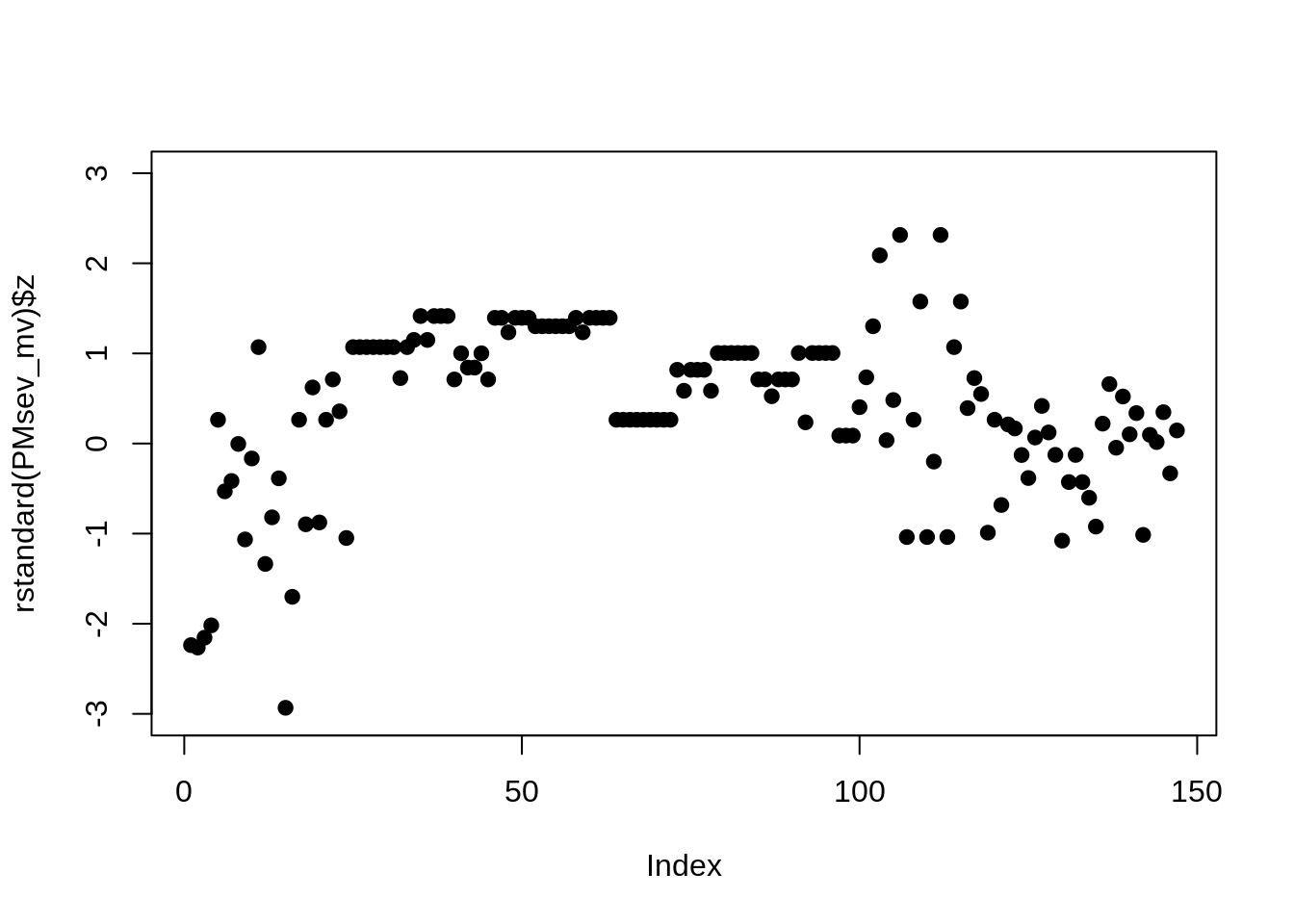
This analysis shows the residual are slightly unbalanced and perhaps an effect of time. Does squaring the response improve the residuals?
PMsev_mvSQ <- rma.mv(
yi = yi_square,
V = vi_square,
mods = ~ spray_management,
method = "ML",
random = list(~ spray_management | trial, ~ 1 | id),
struct = "UN",
control = list(optimizer = "optim"),
data = PM_dat_D
)## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.
summary(PMsev_mvSQ)##
## Multivariate Meta-Analysis Model (k = 147; method: ML)
##
## logLik Deviance AIC BIC AICc
## -532.3560 726.4536 1118.7120 1199.4537 1131.4179
##
## Variance Components:
##
## estim sqrt nlvls fixed factor
## sigma^2 23.3194 4.8290 147 no id
##
## outer factor: trial (nlvls = 22)
## inner factor: spray_management (nlvls = 6)
##
## estim sqrt k.lvl fixed level
## tau^2.1 306.0169 17.4933 37 no control
## tau^2.2 306.3846 17.5038 13 no Early
## tau^2.3 134.8188 11.6111 15 no Late
## tau^2.4 20.9591 4.5781 13 no Late_plus
## tau^2.5 575.9514 23.9990 28 no Recommended
## tau^2.6 891.6918 29.8612 41 no Recommended_plus
##
## rho.cntr rho.Erly rho.Late rho.Lt_p rho.Rcmm rho.Rcm_
## control 1 0.9817 0.9321 1.0000 0.7906 0.8253
## Early 0.9817 1 0.9839 0.0000 0.7816 0.9024
## Late 0.9321 0.9839 1 0.9306 0.7456 0.9440
## Late_plus 1.0000 0.0000 0.9306 1 0.7904 0.8233
## Recommended 0.7906 0.7816 0.7456 0.7904 1 0.8436
## Recommended_plus 0.8253 0.9024 0.9440 0.8233 0.8436 1
## cntr Erly Late Lt_p Rcmm Rcm_
## control - no no no no no
## Early 7 - no yes no no
## Late 9 6 - no no no
## Late_plus 4 0 1 - no no
## Recommended 18 6 9 1 - no
## Recommended_plus 16 6 9 1 16 -
##
## Test for Residual Heterogeneity:
## QE(df = 141) = 30608.5216, p-val < .0001
##
## Test of Moderators (coefficients 2:6):
## QM(df = 5) = 140.8577, p-val < .0001
##
## Model Results:
##
## estimate se zval pval ci.lb
## intrcpt 60.0043 3.8371 15.6378 <.0001 52.4836
## spray_managementEarly -4.0626 1.9081 -2.1292 0.0332 -7.8023
## spray_managementLate -11.9164 2.3694 -5.0292 <.0001 -16.5604
## spray_managementLate_plus -17.4846 3.2357 -5.4037 <.0001 -23.8264
## spray_managementRecommended -17.3870 3.7880 -4.5900 <.0001 -24.8113
## spray_managementRecommended_plus -38.1953 4.6263 -8.2561 <.0001 -47.2628
## ci.ub
## intrcpt 67.5249 ***
## spray_managementEarly -0.3229 *
## spray_managementLate -7.2724 ***
## spray_managementLate_plus -11.1428 ***
## spray_managementRecommended -9.9626 ***
## spray_managementRecommended_plus -29.1278 ***
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 This meta-analysis is more in-line with estimates from the yield meta-analysis.
This meta-analysis is more in-line with estimates from the yield meta-analysis.
Early sprays are not effective at reducing powdery mildew severity and two or more sprays had higher efficacy than single sprays.
Recommended sprays were the most effective spray timing at reducing disease severity.
However square severity is not common when evaluating a response that was obtained on a scale. Log transformed is a more common method of evaluating non-normal disease severity.
Let’s assess the log transformed percent severity.
# remove controls for response ratio meta-analysis
PMsev_mv_log <- rma.mv(
yi = yi_log,
V = yi_log,
mods = ~ spray_management,
method = "ML",
random = list(~ spray_management | trial, ~ 1 | id),
struct = "UN",
control = list(optimizer = "optim"),
data = PM_dat_D
)## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.
summary(PMsev_mv_log)##
## Multivariate Meta-Analysis Model (k = 147; method: ML)
##
## logLik Deviance AIC BIC AICc
## -23.7322 Inf 101.4645 182.2061 114.1703
##
## Variance Components:
##
## estim sqrt nlvls fixed factor
## sigma^2 0.0497 0.2230 147 no id
##
## outer factor: trial (nlvls = 22)
## inner factor: spray_management (nlvls = 6)
##
## estim sqrt k.lvl fixed level
## tau^2.1 0.0207 0.1438 37 no control
## tau^2.2 0.0666 0.2580 13 no Early
## tau^2.3 0.0105 0.1026 15 no Late
## tau^2.4 0.0000 0.0067 13 no Late_plus
## tau^2.5 0.1564 0.3954 28 no Recommended
## tau^2.6 0.2733 0.5228 41 no Recommended_plus
##
## rho.cntr rho.Erly rho.Late rho.Lt_p rho.Rcmm rho.Rcm_
## control 1 0.9272 0.9080 0.9951 0.9649 0.8100
## Early 0.9272 1 0.9988 0.0000 0.7962 0.9707
## Late 0.9080 0.9988 1 0.8622 0.7661 0.9812
## Late_plus 0.9951 0.0000 0.8622 1 0.9861 0.7481
## Recommended 0.9649 0.7962 0.7661 0.9861 1 0.6275
## Recommended_plus 0.8100 0.9707 0.9812 0.7481 0.6275 1
## cntr Erly Late Lt_p Rcmm Rcm_
## control - no no no no no
## Early 7 - no yes no no
## Late 9 6 - no no no
## Late_plus 4 0 1 - no no
## Recommended 18 6 9 1 - no
## Recommended_plus 16 6 9 1 16 -
##
## Test of Moderators (coefficients 2:6):
## QM(df = 5) = 32.1577, p-val < .0001
##
## Model Results:
##
## estimate se zval pval ci.lb
## intrcpt -0.1940 0.0481 -4.0353 <.0001 -0.2883
## spray_managementEarly -0.1571 0.0822 -1.9111 0.0560 -0.3182
## spray_managementLate -0.1017 0.0705 -1.4416 0.1494 -0.2400
## spray_managementLate_plus -0.1391 0.0778 -1.7886 0.0737 -0.2915
## spray_managementRecommended -0.2561 0.0853 -3.0035 0.0027 -0.4232
## spray_managementRecommended_plus -0.5686 0.1165 -4.8810 <.0001 -0.7969
## ci.ub
## intrcpt -0.0998 ***
## spray_managementEarly 0.0040 .
## spray_managementLate 0.0366
## spray_managementLate_plus 0.0133 .
## spray_managementRecommended -0.0890 **
## spray_managementRecommended_plus -0.3403 ***
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
The plot of the residuals show the model is not normally distributed. In addition the estimates don’t reflect what can be observed in the raw data. Log transformed percent severity is therefore no a suitable transformation.
Finally let’s assess the logit transformed percent severity.
# remove controls for response ratio meta-analysis
PMsev_mvPer <- rma.mv(
yi = yi_logit,
V = yi_logit,
mods = ~ spray_management,
method = "ML",
random = list(~ spray_management | trial, ~ 1 | id),
struct = "UN",
control = list(optimizer = "optim"),
data = PM_dat_D
)## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.
summary(PMsev_mvPer)##
## Multivariate Meta-Analysis Model (k = 147; method: ML)
##
## logLik Deviance AIC BIC AICc
## -254.6073 Inf 563.2146 643.9563 575.9205
##
## Variance Components:
##
## estim sqrt nlvls fixed factor
## sigma^2 0.2978 0.5457 147 no id
##
## outer factor: trial (nlvls = 22)
## inner factor: spray_management (nlvls = 6)
##
## estim sqrt k.lvl fixed level
## tau^2.1 4.0250 2.0062 37 no control
## tau^2.2 2.9086 1.7054 13 no Early
## tau^2.3 0.0918 0.3030 15 no Late
## tau^2.4 0.0029 0.0538 13 no Late_plus
## tau^2.5 0.9843 0.9921 28 no Recommended
## tau^2.6 1.2059 1.0982 41 no Recommended_plus
##
## rho.cntr rho.Erly rho.Late rho.Lt_p rho.Rcmm rho.Rcm_
## control 1 0.9992 0.9799 0.9995 0.8628 0.8191
## Early 0.9992 1 0.9869 0.0000 0.8818 0.8407
## Late 0.9799 0.9869 1 0.9733 0.9463 0.9170
## Late_plus 0.9995 0.0000 0.9733 1 0.8467 0.8009
## Recommended 0.8628 0.8818 0.9463 0.8467 1 0.9967
## Recommended_plus 0.8191 0.8407 0.9170 0.8009 0.9967 1
## cntr Erly Late Lt_p Rcmm Rcm_
## control - no no no no no
## Early 7 - no yes no no
## Late 9 6 - no no no
## Late_plus 4 0 1 - no no
## Recommended 18 6 9 1 - no
## Recommended_plus 16 6 9 1 16 -
##
## Test of Moderators (coefficients 2:6):
## QM(df = 5) = 45.9481, p-val < .0001
##
## Model Results:
##
## estimate se zval pval ci.lb
## intrcpt 2.4499 0.5026 4.8748 <.0001 1.4649
## spray_managementEarly -0.4833 0.4791 -1.0089 0.3130 -1.4223
## spray_managementLate -1.2147 0.5648 -2.1509 0.0315 -2.3216
## spray_managementLate_plus -1.5524 0.5787 -2.6825 0.0073 -2.6866
## spray_managementRecommended -1.8393 0.4268 -4.3093 <.0001 -2.6759
## spray_managementRecommended_plus -2.5308 0.4183 -6.0501 <.0001 -3.3506
## ci.ub
## intrcpt 3.4349 ***
## spray_managementEarly 0.4556
## spray_managementLate -0.1078 *
## spray_managementLate_plus -0.4182 **
## spray_managementRecommended -1.0028 ***
## spray_managementRecommended_plus -1.7109 ***
##
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1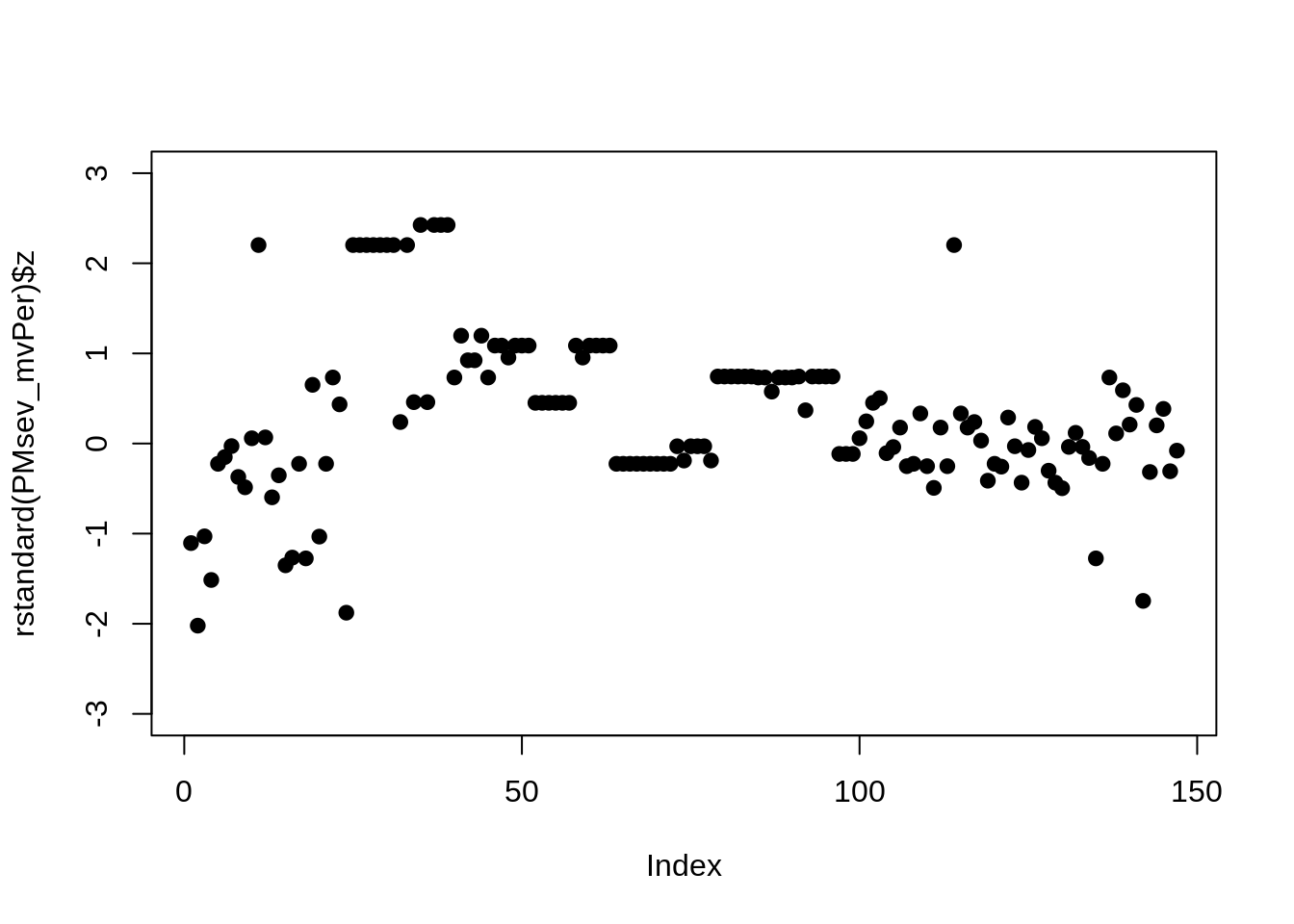
s_analysis <-
lapply(unique(PM_dat_D$trial_ref), function(Trial) {
dat <- filter(PM_dat_D, trial_ref != Trial)
# remove controls for response ratio meta-analysis
rma.mv(
yi = yi_logit,
V = yi_logit,
mods = ~ spray_management,
method = "ML",
random = list( ~ spray_management | trial, ~ 1 | id),
struct = "UN",
control = list(optimizer = "optim"),
data = dat
)
})## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.## Warning: There are outcomes with non-positive sampling variances.## Warning: Negative sampling variances constrained to zero.## Warning: 'V' appears to be not positive definite.## Warning: Some combinations of the levels of the inner factor never occurred.
## Corresponding rho value(s) fixed to 0.
length(s_analysis)## [1] 14
est <- lapply(s_analysis, function(e1) {
data.frame(
treat = rownames(e1$b),
estimates = e1$b,
p = as.numeric(reportP(
e1$pval, P_prefix = FALSE, AsNumeric = TRUE
))
)
})
sensitiviy_df <- data.table::rbindlist(est)
sensitiviy_df$Tnum <- rep(1:14, each = 6)
sensitiviy_df %>%
ggplot(aes(Tnum, estimates, colour = treat)) +
geom_line(size = 1)
sensitiviy_df %>%
ggplot(aes(Tnum, p, colour = treat)) +
geom_line(size = 1) +
scale_y_log10()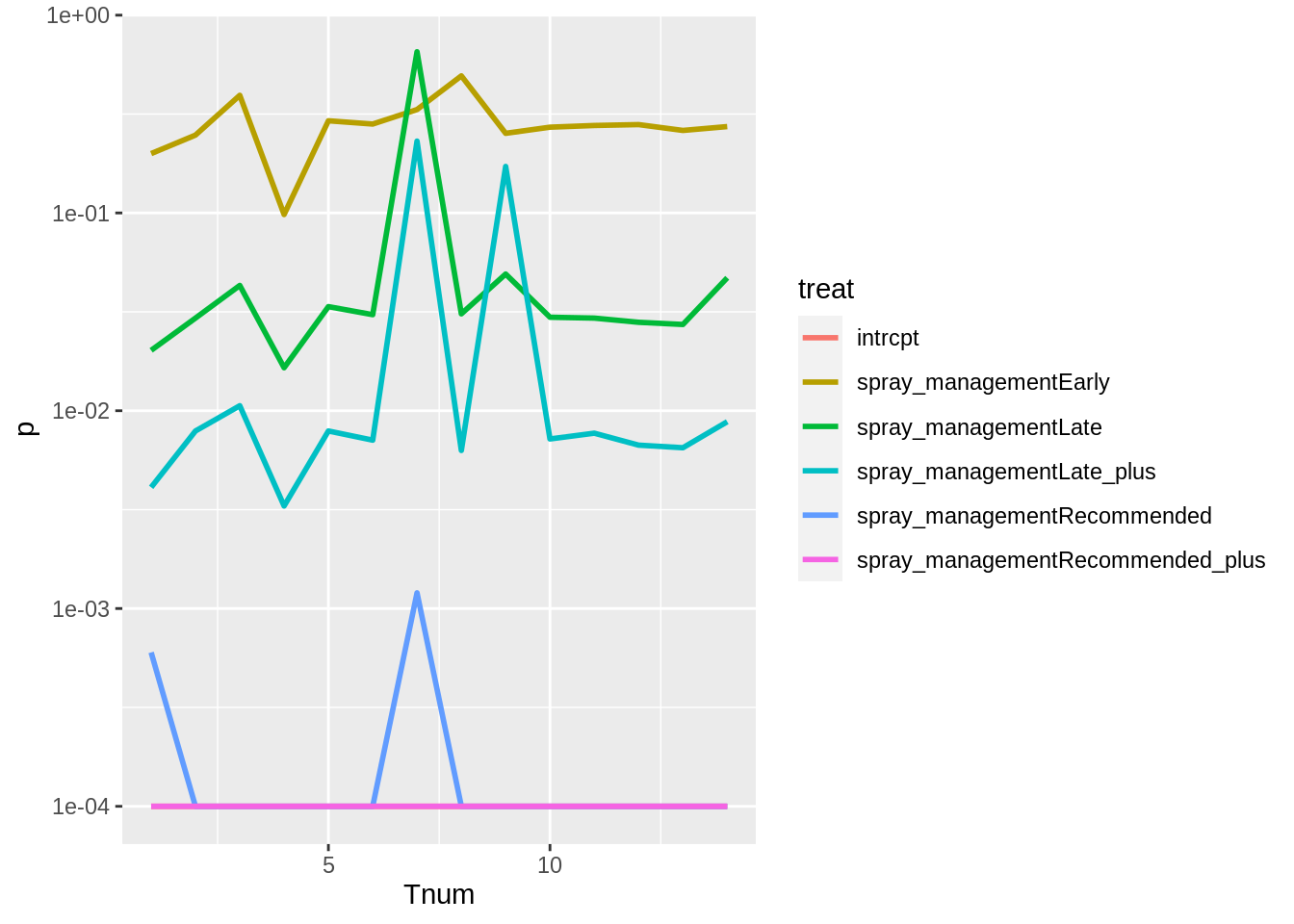
unique(PM_dat_D$trial_ref)[7]## [1] "mung1617/02"
sensitiviy_df %>%
group_by(treat) %>%
summarise(
minEst = min(estimates),
medEst = median(estimates),
maxEst = max(estimates),
minP = min(p),
medP = median(p),
maxP = max(p)
)## # A tibble: 6 × 7
## treat minEst medEst maxEst minP medP maxP
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 intrcpt 1.39 2.51 2.73 0.0001 0.0001 0.0001
## 2 spray_managementEarly -1.08 -0.519 -0.412 0.0982 0.275 0.493
## 3 spray_managementLate -1.54 -1.26 -0.224 0.0165 0.0302 0.653
## 4 spray_managementLate_plus -1.83 -1.61 -0.500 0.0033 0.00745 0.231
## 5 spray_managementRecommended -2.32 -1.89 -1.06 0.0001 0.0001 0.0012
## 6 spray_managementRecommended_plus -3.07 -2.58 -1.82 0.0001 0.0001 0.0001While estimates and P values remained the mostly same when each trial was dropped from the analysis. Estimates changed the most when Trial 1617/02 at Missen Flats in 2017 was ommited from the analysis.
Calculation of I^2.
W <- diag(1 / (PM_dat_D$vi_logit + 0.0001))
X <- model.matrix(PMsev_mvPer)
P <- W - W %*% X %*% solve(t(X) %*% W %*% X) %*% t(X) %*% W
100 * sum(PMsev_mvPer$sigma2) / (sum(PMsev_mvPer$sigma2) + (PMsev_mvPer$k -
PMsev_mvPer$p) / sum(diag(P)))## [1] 99.93606
100 * PMsev_mvPer$sigma2 / (sum(PMsev_mvPer$sigma2) + (PMsev_mvPer$k - PMsev_mvPer$p) /
sum(diag(P)))## [1] 99.93606This indicates that approximate amount of total variance attributed to trials/ clusters is 99%, which is very high. However given that many of the means included in the model contained an accompanying sample variance of zero, this might have been expected.
The estimates and significance from this model better reflect the distribution of raw values. In addition the residuals from this model are more evenly distributed. Let’s back transform the estimates.
backtransform <- function(m, input_only = FALSE, intercept) {
if (input_only == FALSE) {
intercept <- m$b[1]
dat <- data.frame(
estimates = inv_logit(c(intercept, m$b[-1] + intercept)),
StdErr = inv_logit(m$se + intercept) - inv_logit(intercept),
Zval = c(inv_logit(m$z[1]),
inv_logit(m$z[-1] + m$z[1]) - inv_logit(m$z[1])),
Pval = reportP(m$pval, P_prefix = FALSE),
CI_lb = c(inv_logit(m$ci.lb[1]),
inv_logit(m$ci.lb[-1] + intercept)),
CI_ub = c(inv_logit(m$ci.ub[1]),
inv_logit(m$ci.ub[-1] + intercept)),
row.names = colnames(m$G)
)
}
if (input_only == TRUE) {
if (missing(intercept))
stop("'intercept' argument is empty")
return(inv_logit(m + intercept) - inv_logit(intercept))
}
return(dat)
}
backtransform(PMsev_mvPer)## estimates StdErr Zval Pval CI_lb CI_ub
## control 0.9205527 0.02982578 0.99242160 < 0.0001 0.8122794 0.9687768
## Early 0.8772409 0.02870660 -0.01293451 0.313 0.7364486 0.9481188
## Late 0.7747212 0.03267812 -0.05399564 0.0315 0.5320236 0.9123010
## Late_plus 0.7104378 0.03329573 -0.09286533 0.0073 0.4410980 0.8840885
## Recommended 0.6480696 0.02613013 -0.35468527 < 0.0001 0.4437397 0.8095559
## Recommended_plus 0.4797860 0.02569827 -0.75652003 < 0.0001 0.2888949 0.67676775.1.1 Disease severity moderator contrasts
Calculate the treatment contrasts without back-transformation.
source("R/simple_summary.R") #function to provide a table that includes the treatment names in the contrasts
intercept <- PMsev_mvPer$b[1, 1]
z <- PMsev_mvPer$zval[1]
contrast_Ssum <-
simple_summary(summary(glht(PMsev_mvPer, linfct = cbind(
contrMat(rep(1, 6), type = "Tukey")
)), test = adjusted("none")))
contrast_Ssum[6:15, ] %>%
flextable() %>%
autofit()contrast |
coefficients |
StdErr |
Zvalue |
pvals |
sig |
Late - Early |
-0.7314 |
0.6346 |
-1.1525 |
0.2491 |
|
Late_plus - Early |
-1.0690 |
0.6475 |
-1.6510 |
0.0987 |
|
Recommended - Early |
-1.3560 |
0.5373 |
-2.5236 |
0.0116 |
* |
Recommended_plus - Early |
-2.0475 |
0.5263 |
-3.8906 |
< 0.0001 |
*** |
Late_plus - Late |
-0.3377 |
0.4608 |
-0.7328 |
0.4637 |
|
Recommended - Late |
-0.6246 |
0.4244 |
-1.4718 |
0.1411 |
|
Recommended_plus - Late |
-1.3161 |
0.4175 |
-3.1524 |
0.0016 |
** |
Recommended - Late_plus |
-0.2869 |
0.4171 |
-0.6880 |
0.4914 |
|
Recommended_plus - Late_plus |
-0.9784 |
0.4131 |
-2.3682 |
0.0179 |
* |
Recommended_plus - Recommended |
-0.6915 |
0.2400 |
-2.8807 |
0.004 |
** |
par(mar = c(5, 13, 4, 2) + 0.1)
plot(glht(PMsev_mvPer, linfct = cbind(contrMat(rep(
1, 6
), type = "Tukey"))), yaxt = 'n')
axis(
2,
at = seq_along(contrast_Ssum$contrast),
labels = rev(contrast_Ssum$contrast),
las = 2,
cex.axis = 0.8
)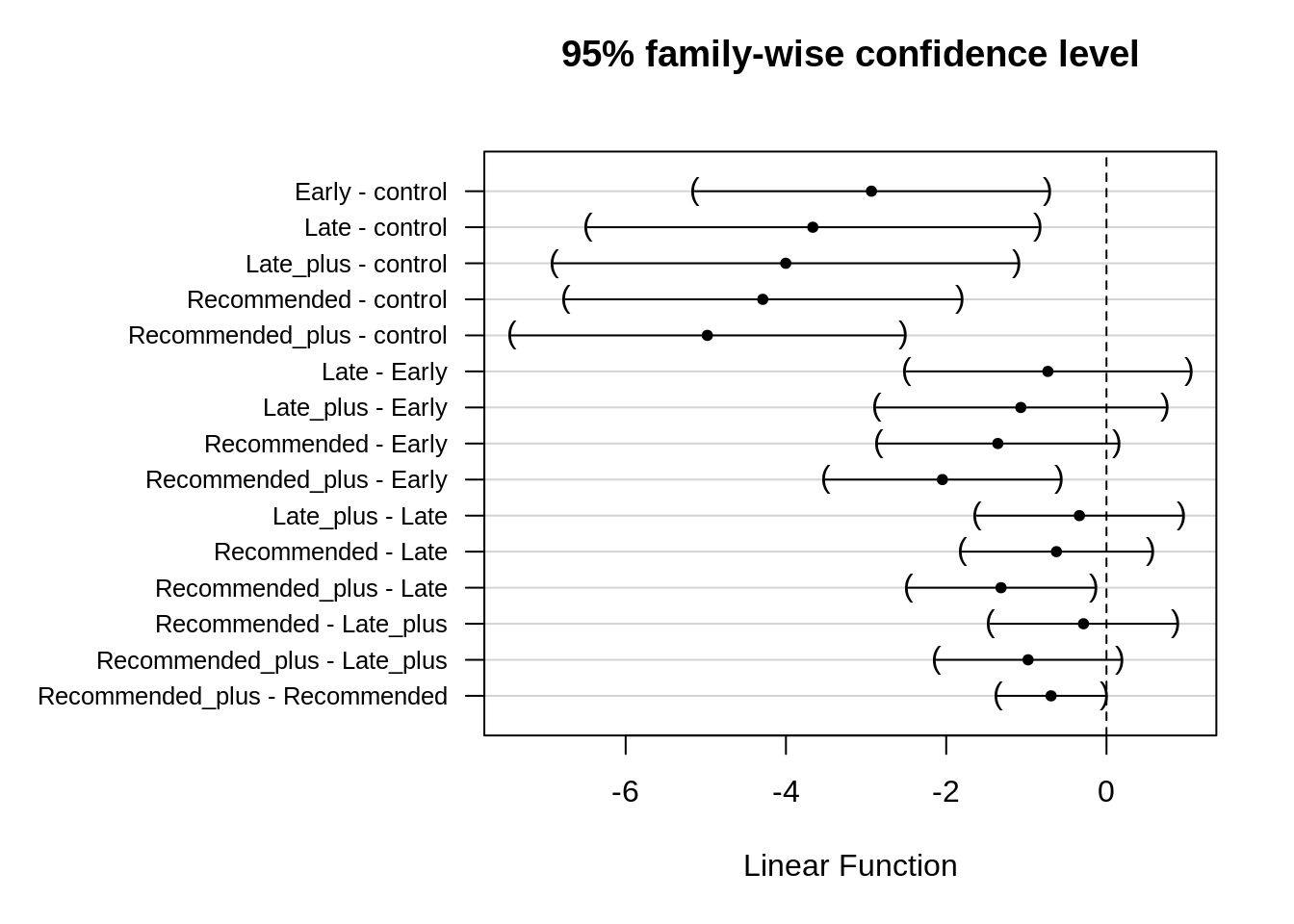
Recommended_plus limited disease severity significantly better than any other spray treatment.
Early sprays were not effective in decreasing disease severity.
Two spray treatments were significantly better than a single application, but the timing of the fungicide application was more critical.
5.1.2 Meta-analysis summary table
# obtain number of treatments included in each moderator variable
k5 <-
as.data.frame(table(PM_dat_D$spray_management)) %>%
filter(Freq != 0) %>%
pull(Freq)
k6 <-
as.data.frame(table(PM_dat_D$trial_ref, PM_dat_D$spray_management)) %>%
filter(Freq != 0) %>%
group_by(Var2) %>%
summarise(n()) %>%
pull()
# create data.frame of percent disease severity estimates
results_mv <- cbind(data.frame(
Moderator = c(
"Intercept / No Spray control",
"Early",
"Late",
"Late+",
"Recommended",
"Recommended+"
),
N = k5,
k = k6
),
backtransform(PMsev_mvPer))
# Calculate mean differences
Intcpt <- results_mv$estimates[1]
results_mv <-
results_mv %>%
mutate(
estimates = estimates - Intcpt,
CI_lb = CI_lb - Intcpt,
CI_ub = CI_lb - Intcpt
)
# round the numbers
results_mv[, c(4:6, 8:9)] <- round(results_mv[, c(4:6, 8:9)], 4)
# rename colnames to give table headings
colnames(results_mv)[c(1:9)] <-
c("Moderator",
"N",
"k",
"mu",
"se",
"Z",
"P",
"CI_{L}",
"CI_{U}")
disease_estimates_table <-
results_mv[c(2, 5, 6, 3, 4), -6] %>%
flextable() %>%
align(j = 2:8, align = "center", part = "all") %>%
fontsize(size = 8, part = "body") %>%
fontsize(size = 10, part = "header") %>%
italic(italic = TRUE, part = "header") %>%
set_caption(
"Table 3: Estimated powdery mildew severity mean difference for each spray schedule treatment to the unsprayed control treatments. Meta-analysis estimates were back transformed using an inverse logit. Data were obtained from the grey literature reports of (k) field trials undertaken in Eastern Australia. P values indicate statistical significance in comparison to the intercept."
) %>%
footnote(
i = 1,
j = c(2:4, 6:8),
value = as_paragraph(
c(
"Number of treatment means categorised to each spray schedule",
"Number of trials with the respective spray schedule",
"Estimated mean disease severity",
"Indicates the significance between each respective spray schedule and the no spray control (intercept)",
"Lower range of the 95% confidence interval",
"Upper range of the 95% confidence interval"
)
),
ref_symbols = letters[c(1:6)],
part = "header",
inline = TRUE
) %>%
hline_top(part = "all",
border = officer::fp_border(color = "black", width = 2)) %>%
width(j = 1, width = 1) %>%
width(j = 2, width = 0.3) %>%
width(j = 3, width = 0.3) %>%
width(j = 4:8, width = 0.6)
disease_estimates_tableModerator |
Na |
kb |
muc |
se |
Pd |
CI_{L}e |
CI_{U}f |
Early |
13 |
3 |
-0.0433 |
0.0287 |
0.313 |
-0.1841 |
-1.1047 |
Recommended |
28 |
12 |
-0.2725 |
0.0261 |
< 0.0001 |
-0.4768 |
-1.3974 |
Recommended+ |
41 |
11 |
-0.4408 |
0.0257 |
< 0.0001 |
-0.6317 |
-1.5522 |
Late |
15 |
5 |
-0.1458 |
0.0327 |
0.0315 |
-0.3885 |
-1.3091 |
Late+ |
13 |
2 |
-0.2101 |
0.0333 |
0.0073 |
-0.4795 |
-1.4000 |
aNumber of treatment means categorised to each spray schedule; bNumber of trials with the respective spray schedule; cEstimated mean disease severity; dIndicates the significance between each respective spray schedule and the no spray control (intercept); eLower range of the 95% confidence interval; fUpper range of the 95% confidence interval | |||||||
Kelly, Lisa, Jo White, Murray Sharman, Hugh Brier, Liz Williams, Raechelle Grams, Duncan Weir, Alan Mckay, and Adam H. Sparks. 2017. “Mungbean and sorghum disease update - GRDC.” https://grdc.com.au/resources-and-publications/grdc-update-papers/tab-content/grdc-update-papers/2017/07/mungbean-and-sorghum-disease-update.
Madden, L. V., H. P. Piepho, and P. A. Paul. 2016. “Statistical Models and Methods for Network Meta-Analysis.” Phytopathology 106 (8): 792–806. https://doi.org/10.1094/PHYTO-12-15-0342-RVW.
Thompson, Sue. 2016. “Mungbeans Vs Fungus: Two Sprays for Optimum Control - Grains Research and Development Corporation.” Ground Cover, September. https://grdc.com.au/resources-and-publications/groundcover/ground-cover-issue-124-septemberoctober-2016/mungbeans-vs-fungus-two-sprays-for-optimum-control.
Viechtbauer, Wolfgang. 2010. “Conducting Meta-Analyses in R with the metafor Package.” Journal of Statistical Software 36 (3): 1–48. https://doi.org/10.18637/jss.v036.i03.